Gene
KWMTBOMO11771 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000201
Annotation
proteasome_beta_subunit_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.51 Mitochondrial Reliability : 1.226
Sequence
CDS
ATGTTGAGCGTTAACGAGAACTTTCCTGAATATGCTGTGCCTGGTGCGAAACAGCATCGTTTCGAGCCCTACGCGGACAACGGAGGTAGTATTGTAGCCATCGCCGGCGATGATTATGCCGTTATCGGAGCAGACACTCGTCTCAGCACTGGATTTTCCATATATACCAGAGACCAAAAAAAGCTGTTCAAATTATCTGAGAGTACAGTATTGGGAGCGACTGGCTGCTGGTGTGACACATTAACCTTGACGCGTCTTCTACAAGCGAGGATGCAGATGTACGAACACGAGCATAACAAATCTATGACCACTCCGGCAGTCGCACAGATGCTCTCAACTATGTTGTATTACAAACGATTTTTCCCATACTACGTATCCAACGTGTTGGCGGGTTTGGACAGCGACGGTAAGGGATGTGTATATAGTTACGACCCTATCGGACACTGTGCGCGCCATAACTTCCGCGCTGGTGGTTCCGCTGCTGCACAGCTGCAACCGCTCCTCGACAATCAAATTGGCTTAAAGAATATGCAGAATGTAACAGAAGCTCCTTTGCCTAGGGAAAAGGCCCTTGCTCTTCTGAAGGATGTGTTCATCAGTGCTGCAGAACGTGACATTTACACTGGAGACTCCATCTACATTCTCATCATCACTGCAAGTGGTATTCAAGAAGAGAAATTTGAACTTCGCAAAGATTAA
Protein
MLSVNENFPEYAVPGAKQHRFEPYADNGGSIVAIAGDDYAVIGADTRLSTGFSIYTRDQKKLFKLSESTVLGATGCWCDTLTLTRLLQARMQMYEHEHNKSMTTPAVAQMLSTMLYYKRFFPYYVSNVLAGLDSDGKGCVYSYDPIGHCARHNFRAGGSAAAQLQPLLDNQIGLKNMQNVTEAPLPREKALALLKDVFISAAERDIYTGDSIYILIITASGIQEEKFELRKD
Summary
Uniprot
Q1HQ18
A0A2H1W3L5
A0A212EJX4
A0A2A4IUS8
I4DMH1
A0A194QZI2
+ More
B6ZAY6 S4PGV0 I4DJP0 A0A0L7KUS6 A0A194PM38 A0A1E1WL98 A0A1W4WLK5 A0A2P8YPU4 D6WD83 A0A2D1QUD1 N6TDD9 A0A1Y1NFB8 A0A3L8DJD5 A0A2A3ENL7 A0A088A9Q2 A0A0L7QX41 E9IJS5 E2AV04 A0A0N0BD07 A0A2J7RF54 E2B518 A0A067QEY5 A0A195B8T3 F4X0J5 A0A158ND25 A0A151IDY1 A0A151XG90 A0A0C9QBN9 A0A1B1ZGH5 A0A195DUB4 A0A023EKH7 Q16XT2 A0A182N1W0 A0A084WDX4 A0A182J7S9 A0A0T6B1G7 A0A182YQQ9 A0A151IDU8 A0A182QBF3 B0WAD5 A0A182REP1 K7IW45 A0A2R5LC90 A0A1Q3FCX2 A0A182JV84 A0A232EMK4 A0A182SXQ6 A0A182VFU2 A0A182KNL1 A0A182XJH4 Q7Q7W2 A0A182HQP2 A0A182PUQ2 T1J791 A0A182ULG6 A0A182LSN9 A0A182WM44 A0A2M4AH82 A0A2M4AH89 A0A2M3ZHI7 A0A1Z5KX81 A0A1B6C5V8 W5J9I3 A0A023FK16 A0A293N2G5 A0A023FUQ6 A0A023GJW8 G3MRB6 A0A1I8NDR4 A0A1E1X2Y9 A0A0L0BUP5 A0A182FL90 W8BYK0 A0A034WTT2 A0A0K8VUF8 A0A154NYQ9 B4L9S3 A0A0M4EKH7 T1EAP6 A0A2M4BZK1 B4LCY3 E0W1R5 A0A224YRQ2 A0A0A1X298 A0A0P4VNN7 R4G558 A0A131XK59 A0A131YJ78 L7M7J5 A0A1L8EE81 A0A1I8P9I5 B4J0F1 A0A164Q8S6 A0A0K8R8H6
B6ZAY6 S4PGV0 I4DJP0 A0A0L7KUS6 A0A194PM38 A0A1E1WL98 A0A1W4WLK5 A0A2P8YPU4 D6WD83 A0A2D1QUD1 N6TDD9 A0A1Y1NFB8 A0A3L8DJD5 A0A2A3ENL7 A0A088A9Q2 A0A0L7QX41 E9IJS5 E2AV04 A0A0N0BD07 A0A2J7RF54 E2B518 A0A067QEY5 A0A195B8T3 F4X0J5 A0A158ND25 A0A151IDY1 A0A151XG90 A0A0C9QBN9 A0A1B1ZGH5 A0A195DUB4 A0A023EKH7 Q16XT2 A0A182N1W0 A0A084WDX4 A0A182J7S9 A0A0T6B1G7 A0A182YQQ9 A0A151IDU8 A0A182QBF3 B0WAD5 A0A182REP1 K7IW45 A0A2R5LC90 A0A1Q3FCX2 A0A182JV84 A0A232EMK4 A0A182SXQ6 A0A182VFU2 A0A182KNL1 A0A182XJH4 Q7Q7W2 A0A182HQP2 A0A182PUQ2 T1J791 A0A182ULG6 A0A182LSN9 A0A182WM44 A0A2M4AH82 A0A2M4AH89 A0A2M3ZHI7 A0A1Z5KX81 A0A1B6C5V8 W5J9I3 A0A023FK16 A0A293N2G5 A0A023FUQ6 A0A023GJW8 G3MRB6 A0A1I8NDR4 A0A1E1X2Y9 A0A0L0BUP5 A0A182FL90 W8BYK0 A0A034WTT2 A0A0K8VUF8 A0A154NYQ9 B4L9S3 A0A0M4EKH7 T1EAP6 A0A2M4BZK1 B4LCY3 E0W1R5 A0A224YRQ2 A0A0A1X298 A0A0P4VNN7 R4G558 A0A131XK59 A0A131YJ78 L7M7J5 A0A1L8EE81 A0A1I8P9I5 B4J0F1 A0A164Q8S6 A0A0K8R8H6
Pubmed
19121390
22118469
22651552
26354079
28756777
23622113
+ More
26227816 29403074 18362917 19820115 23537049 28004739 30249741 21282665 20798317 24845553 21719571 21347285 27464714 24945155 26483478 17510324 24438588 25244985 20075255 28648823 20966253 12364791 14747013 17210077 28528879 20920257 23761445 22216098 25315136 28503490 26108605 24495485 25348373 17994087 20566863 28797301 25830018 27129103 28049606 26830274 25576852
26227816 29403074 18362917 19820115 23537049 28004739 30249741 21282665 20798317 24845553 21719571 21347285 27464714 24945155 26483478 17510324 24438588 25244985 20075255 28648823 20966253 12364791 14747013 17210077 28528879 20920257 23761445 22216098 25315136 28503490 26108605 24495485 25348373 17994087 20566863 28797301 25830018 27129103 28049606 26830274 25576852
EMBL
BABH01040705
DQ443234
ABF51323.1
ODYU01006118
SOQ47680.1
AGBW02014361
+ More
OWR41803.1 NWSH01006778 PCG63254.1 PCG63256.1 AK402489 BAM19111.1 KQ460949 KPJ10385.1 FJ378903 KZ149915 ACJ13433.1 PZC77978.1 GAIX01006055 JAA86505.1 AK401508 BAM18130.1 JTDY01005418 KOB67022.1 KQ459598 KPI94501.1 GDQN01003418 JAT87636.1 PYGN01000445 PSN46224.1 KQ971321 EEZ99493.1 KY285073 ATP16176.1 APGK01034609 KB740914 KB632352 ENN78384.1 ERL93261.1 GEZM01004165 JAV96449.1 QOIP01000007 RLU20507.1 KZ288212 PBC32842.1 KQ414705 KOC63109.1 GL763883 EFZ19173.1 GL442974 EFN62722.1 KQ435891 KOX69414.1 NEVH01004413 PNF39466.1 GL445685 EFN89226.1 KK853682 KDQ97904.1 KQ976558 KYM80650.1 GL888498 EGI59987.1 ADTU01012187 KQ977906 KYM98870.1 KQ982173 KYQ59414.1 GBYB01000664 GBYB01012844 JAG70431.1 JAG82611.1 KU756279 ANX99822.1 KQ980341 KYN16473.1 JXUM01057612 JXUM01086775 GAPW01003956 KQ563590 KQ561963 JAC09642.1 KXJ73624.1 KXJ77046.1 CH477533 EAT39423.1 ATLV01023078 KE525340 KFB48418.1 LJIG01016246 KRT81178.1 KYM98866.1 AXCN02000973 DS231870 EDS41025.1 GGLE01003004 MBY07130.1 GFDL01009686 JAV25359.1 NNAY01003352 OXU19567.1 AAAB01008948 EAA10482.4 APCN01004672 JH431916 AXCM01006066 GGFK01006822 MBW40143.1 GGFK01006823 MBW40144.1 GGFM01007263 MBW28014.1 GFJQ02007253 JAV99716.1 GEDC01028412 JAS08886.1 ADMH02002077 ETN59520.1 GBBK01003524 JAC20958.1 GFWV01022536 MAA47263.1 GBBL01001804 JAC25516.1 GBBM01002368 JAC33050.1 JO844417 AEO36034.1 GFAC01005586 JAT93602.1 JRES01001437 KNC22929.1 GAMC01002143 JAC04413.1 GAKP01001235 JAC57717.1 GDHF01017320 GDHF01009792 GDHF01002425 JAI34994.1 JAI42522.1 JAI49889.1 KQ434783 KZC04809.1 CH933816 EDW17121.1 CP012525 ALC44688.1 GAMD01000227 JAB01364.1 GGFJ01009017 MBW58158.1 CH940647 EDW68844.1 DS235873 EEB19647.1 GFPF01005508 MAA16654.1 GBXI01009281 JAD05011.1 GDKW01002618 JAI53977.1 ACPB03014465 GAHY01000191 JAA77319.1 GEFH01001829 JAP66752.1 GEDV01010576 JAP77981.1 GACK01005921 JAA59113.1 GFDG01001791 JAV17008.1 CH916366 EDV96787.1 LRGB01002451 KZS07533.1 GADI01006650 JAA67158.1
OWR41803.1 NWSH01006778 PCG63254.1 PCG63256.1 AK402489 BAM19111.1 KQ460949 KPJ10385.1 FJ378903 KZ149915 ACJ13433.1 PZC77978.1 GAIX01006055 JAA86505.1 AK401508 BAM18130.1 JTDY01005418 KOB67022.1 KQ459598 KPI94501.1 GDQN01003418 JAT87636.1 PYGN01000445 PSN46224.1 KQ971321 EEZ99493.1 KY285073 ATP16176.1 APGK01034609 KB740914 KB632352 ENN78384.1 ERL93261.1 GEZM01004165 JAV96449.1 QOIP01000007 RLU20507.1 KZ288212 PBC32842.1 KQ414705 KOC63109.1 GL763883 EFZ19173.1 GL442974 EFN62722.1 KQ435891 KOX69414.1 NEVH01004413 PNF39466.1 GL445685 EFN89226.1 KK853682 KDQ97904.1 KQ976558 KYM80650.1 GL888498 EGI59987.1 ADTU01012187 KQ977906 KYM98870.1 KQ982173 KYQ59414.1 GBYB01000664 GBYB01012844 JAG70431.1 JAG82611.1 KU756279 ANX99822.1 KQ980341 KYN16473.1 JXUM01057612 JXUM01086775 GAPW01003956 KQ563590 KQ561963 JAC09642.1 KXJ73624.1 KXJ77046.1 CH477533 EAT39423.1 ATLV01023078 KE525340 KFB48418.1 LJIG01016246 KRT81178.1 KYM98866.1 AXCN02000973 DS231870 EDS41025.1 GGLE01003004 MBY07130.1 GFDL01009686 JAV25359.1 NNAY01003352 OXU19567.1 AAAB01008948 EAA10482.4 APCN01004672 JH431916 AXCM01006066 GGFK01006822 MBW40143.1 GGFK01006823 MBW40144.1 GGFM01007263 MBW28014.1 GFJQ02007253 JAV99716.1 GEDC01028412 JAS08886.1 ADMH02002077 ETN59520.1 GBBK01003524 JAC20958.1 GFWV01022536 MAA47263.1 GBBL01001804 JAC25516.1 GBBM01002368 JAC33050.1 JO844417 AEO36034.1 GFAC01005586 JAT93602.1 JRES01001437 KNC22929.1 GAMC01002143 JAC04413.1 GAKP01001235 JAC57717.1 GDHF01017320 GDHF01009792 GDHF01002425 JAI34994.1 JAI42522.1 JAI49889.1 KQ434783 KZC04809.1 CH933816 EDW17121.1 CP012525 ALC44688.1 GAMD01000227 JAB01364.1 GGFJ01009017 MBW58158.1 CH940647 EDW68844.1 DS235873 EEB19647.1 GFPF01005508 MAA16654.1 GBXI01009281 JAD05011.1 GDKW01002618 JAI53977.1 ACPB03014465 GAHY01000191 JAA77319.1 GEFH01001829 JAP66752.1 GEDV01010576 JAP77981.1 GACK01005921 JAA59113.1 GFDG01001791 JAV17008.1 CH916366 EDV96787.1 LRGB01002451 KZS07533.1 GADI01006650 JAA67158.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000037510
UP000053268
+ More
UP000192223 UP000245037 UP000007266 UP000019118 UP000030742 UP000279307 UP000242457 UP000005203 UP000053825 UP000000311 UP000053105 UP000235965 UP000008237 UP000027135 UP000078540 UP000007755 UP000005205 UP000078542 UP000075809 UP000078492 UP000069940 UP000249989 UP000008820 UP000075884 UP000030765 UP000075880 UP000076408 UP000075886 UP000002320 UP000075900 UP000002358 UP000075881 UP000215335 UP000075901 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075885 UP000075902 UP000075883 UP000075920 UP000000673 UP000095301 UP000037069 UP000069272 UP000076502 UP000009192 UP000092553 UP000008792 UP000009046 UP000015103 UP000095300 UP000001070 UP000076858
UP000192223 UP000245037 UP000007266 UP000019118 UP000030742 UP000279307 UP000242457 UP000005203 UP000053825 UP000000311 UP000053105 UP000235965 UP000008237 UP000027135 UP000078540 UP000007755 UP000005205 UP000078542 UP000075809 UP000078492 UP000069940 UP000249989 UP000008820 UP000075884 UP000030765 UP000075880 UP000076408 UP000075886 UP000002320 UP000075900 UP000002358 UP000075881 UP000215335 UP000075901 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075885 UP000075902 UP000075883 UP000075920 UP000000673 UP000095301 UP000037069 UP000069272 UP000076502 UP000009192 UP000092553 UP000008792 UP000009046 UP000015103 UP000095300 UP000001070 UP000076858
PRIDE
Pfam
PF00227 Proteasome
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
Q1HQ18
A0A2H1W3L5
A0A212EJX4
A0A2A4IUS8
I4DMH1
A0A194QZI2
+ More
B6ZAY6 S4PGV0 I4DJP0 A0A0L7KUS6 A0A194PM38 A0A1E1WL98 A0A1W4WLK5 A0A2P8YPU4 D6WD83 A0A2D1QUD1 N6TDD9 A0A1Y1NFB8 A0A3L8DJD5 A0A2A3ENL7 A0A088A9Q2 A0A0L7QX41 E9IJS5 E2AV04 A0A0N0BD07 A0A2J7RF54 E2B518 A0A067QEY5 A0A195B8T3 F4X0J5 A0A158ND25 A0A151IDY1 A0A151XG90 A0A0C9QBN9 A0A1B1ZGH5 A0A195DUB4 A0A023EKH7 Q16XT2 A0A182N1W0 A0A084WDX4 A0A182J7S9 A0A0T6B1G7 A0A182YQQ9 A0A151IDU8 A0A182QBF3 B0WAD5 A0A182REP1 K7IW45 A0A2R5LC90 A0A1Q3FCX2 A0A182JV84 A0A232EMK4 A0A182SXQ6 A0A182VFU2 A0A182KNL1 A0A182XJH4 Q7Q7W2 A0A182HQP2 A0A182PUQ2 T1J791 A0A182ULG6 A0A182LSN9 A0A182WM44 A0A2M4AH82 A0A2M4AH89 A0A2M3ZHI7 A0A1Z5KX81 A0A1B6C5V8 W5J9I3 A0A023FK16 A0A293N2G5 A0A023FUQ6 A0A023GJW8 G3MRB6 A0A1I8NDR4 A0A1E1X2Y9 A0A0L0BUP5 A0A182FL90 W8BYK0 A0A034WTT2 A0A0K8VUF8 A0A154NYQ9 B4L9S3 A0A0M4EKH7 T1EAP6 A0A2M4BZK1 B4LCY3 E0W1R5 A0A224YRQ2 A0A0A1X298 A0A0P4VNN7 R4G558 A0A131XK59 A0A131YJ78 L7M7J5 A0A1L8EE81 A0A1I8P9I5 B4J0F1 A0A164Q8S6 A0A0K8R8H6
B6ZAY6 S4PGV0 I4DJP0 A0A0L7KUS6 A0A194PM38 A0A1E1WL98 A0A1W4WLK5 A0A2P8YPU4 D6WD83 A0A2D1QUD1 N6TDD9 A0A1Y1NFB8 A0A3L8DJD5 A0A2A3ENL7 A0A088A9Q2 A0A0L7QX41 E9IJS5 E2AV04 A0A0N0BD07 A0A2J7RF54 E2B518 A0A067QEY5 A0A195B8T3 F4X0J5 A0A158ND25 A0A151IDY1 A0A151XG90 A0A0C9QBN9 A0A1B1ZGH5 A0A195DUB4 A0A023EKH7 Q16XT2 A0A182N1W0 A0A084WDX4 A0A182J7S9 A0A0T6B1G7 A0A182YQQ9 A0A151IDU8 A0A182QBF3 B0WAD5 A0A182REP1 K7IW45 A0A2R5LC90 A0A1Q3FCX2 A0A182JV84 A0A232EMK4 A0A182SXQ6 A0A182VFU2 A0A182KNL1 A0A182XJH4 Q7Q7W2 A0A182HQP2 A0A182PUQ2 T1J791 A0A182ULG6 A0A182LSN9 A0A182WM44 A0A2M4AH82 A0A2M4AH89 A0A2M3ZHI7 A0A1Z5KX81 A0A1B6C5V8 W5J9I3 A0A023FK16 A0A293N2G5 A0A023FUQ6 A0A023GJW8 G3MRB6 A0A1I8NDR4 A0A1E1X2Y9 A0A0L0BUP5 A0A182FL90 W8BYK0 A0A034WTT2 A0A0K8VUF8 A0A154NYQ9 B4L9S3 A0A0M4EKH7 T1EAP6 A0A2M4BZK1 B4LCY3 E0W1R5 A0A224YRQ2 A0A0A1X298 A0A0P4VNN7 R4G558 A0A131XK59 A0A131YJ78 L7M7J5 A0A1L8EE81 A0A1I8P9I5 B4J0F1 A0A164Q8S6 A0A0K8R8H6
PDB
6MSK
E-value=2.74067e-77,
Score=732
Ontologies
KEGG
GO
PANTHER
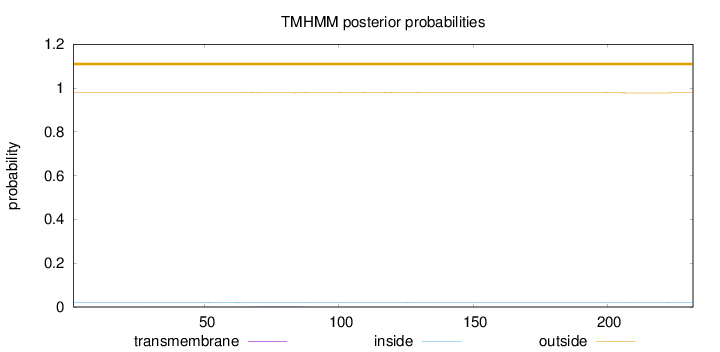
Topology
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09703
Exp number, first 60 AAs:
0.00598
Total prob of N-in:
0.02015
outside
1 - 232
Population Genetic Test Statistics
Pi
13.906861
Theta
10.746567
Tajima's D
0.863702
CLR
1.007115
CSRT
0.634618269086546
Interpretation
Uncertain
