Gene
KWMTBOMO11770
Annotation
hypothetical_protein_M513_10814?_partial_[Trichuris_suis]
Location in the cell
Nuclear Reliability : 3.808
Sequence
CDS
ATGCAAGGCATTGATAAAACGGACCCACCCCCCACAGACCCATTATTTCCTCAACCTACCTCAGTATGCAGAGTAGCCATAAAAATTCCTCCCTTCTGGCCTGATCGGCCTGCAATTTGGTTTGCTCAGGTAGAGGCACAATTCTCGATTTCAGGCATCTCCGCTGACCTGACTAAATTTAACTACGTGATTGCTCAGTTAGATACCCGAGTTATTGGGGAGGTAGAGGACATTATTCTCCAACCACCACCTGAGGACAAATATGGTCGCCTCAAATCCGAGTTGATCCGTAGGTTGTCCACGTCTGAGGAGCAACGTGTCAGGCAACTTGTCAGTGATGTTGAGTTGGGTGACCGCCGTCCTTCGCAGTTTTTACGTTATCTTAGATCATTGGCAGGAAAAACCCTCACTGATGAAAATTTGCTTCGCCAGCTATGGATGCGTCGCCTCCCACAACATCTCCAAGCAATTTTGGCCGCCCAATCCGAACTCTCGCTGGACAAAGTCGCTGAGCTAGCGGACAAAATTTTGGAAGTTTCGCCTGGTATGGTCGCACCTCATTCATCTGCTGTTTTCTCAGCAAGTACGTCTGCCGTCTCACCCCCTTCGTTAGAAGATATCACACGGCGTCTGGATGAGGTCACGCGTCAGCTTGCTGTGCTTACTGCAGAGCGCGGTAGAAATAGGACCCGGGATTTCTCAAACCAGCGACGACGCTCACAAACGCCTGGTAAGCTTCAAATGTGCTTTTACCATAGGAAATTTGATACTAAAGCTGTTAAGTGTATTCAACCATGTACCTGGACGCCGCCGCCATCTTCGGAAAACGACAAAAGCAGTCAATGA
Protein
MQGIDKTDPPPTDPLFPQPTSVCRVAIKIPPFWPDRPAIWFAQVEAQFSISGISADLTKFNYVIAQLDTRVIGEVEDIILQPPPEDKYGRLKSELIRRLSTSEEQRVRQLVSDVELGDRRPSQFLRYLRSLAGKTLTDENLLRQLWMRRLPQHLQAILAAQSELSLDKVAELADKILEVSPGMVAPHSSAVFSASTSAVSPPSLEDITRRLDEVTRQLAVLTAERGRNRTRDFSNQRRRSQTPGKLQMCFYHRKFDTKAVKCIQPCTWTPPPSSENDKSSQ
Summary
Uniprot
A0A085LTM6
A0A085N4U2
A0A085NBV3
A0A0J7K6V3
A0A0J7K6I6
A0A151IZ47
+ More
A0A151ITG2 A0A0J7KD10 A0A0J7K7T5 A0A2J7RQK2 A0A1Y1MXV9 A0A2A4J8M7 A0A151JCH0 A0A1W4XBP5 A0A0L7QJB5 A0A0L7QK36 A0A2J7PLR6 A0A2J7QTC8 A0A0J7K0A7 A0A151K0A8 T1IE13 A0A0J7K6V1 A0A2J7QJD3 A0A026VRX8 A0A2G8L3A3 A0A2J7RGP3 E2BBH0 A0A2J7QH56 A0A154P674 A0A2H1WXE7 A0A2A4K2H4 A0A154NZG0 A0A2J7PML8 A0A2G8LNJ3 A0A1Y1MA44 A0A1E1W488 A0A2J7PCS9 A0A1S3CY78 A0A131YTY1 A0A151K2U9 A0A151JF00 A0A2A4JHF4 W4YWZ1 A0A1I8Q981 A0A0N5DP72 A0A0P8Y533 A0A0L7QMU1 A0A2L2YSA7 A0A0P8ZL89 W8B3F8 A0A293M3N6 A0A085LQ61 A0A0L7QJ97 A0A0L7QJ51 A0A131YM78 A0A139WA14 D7GYF3 A0A2G8JL30 D7EJX5 A0A3L8DMT5 A0A154PMS5 A0A1Y1NAW7 A0A182G9J3 A0A182H518 A0A151JBY5 D6WTN9 A0A0A9YNN7 A0A139W8G7 A0A293M349 A0A085N3N9 A0A182HB40 A0A2L2YTR5 A0A085NTM8 A0A2J7PD24 A0A151IIJ9 A0A026VX87 A0A0J7NAK0 A0A182H3L9 A0A154PT26 A0A0L8GMC8
A0A151ITG2 A0A0J7KD10 A0A0J7K7T5 A0A2J7RQK2 A0A1Y1MXV9 A0A2A4J8M7 A0A151JCH0 A0A1W4XBP5 A0A0L7QJB5 A0A0L7QK36 A0A2J7PLR6 A0A2J7QTC8 A0A0J7K0A7 A0A151K0A8 T1IE13 A0A0J7K6V1 A0A2J7QJD3 A0A026VRX8 A0A2G8L3A3 A0A2J7RGP3 E2BBH0 A0A2J7QH56 A0A154P674 A0A2H1WXE7 A0A2A4K2H4 A0A154NZG0 A0A2J7PML8 A0A2G8LNJ3 A0A1Y1MA44 A0A1E1W488 A0A2J7PCS9 A0A1S3CY78 A0A131YTY1 A0A151K2U9 A0A151JF00 A0A2A4JHF4 W4YWZ1 A0A1I8Q981 A0A0N5DP72 A0A0P8Y533 A0A0L7QMU1 A0A2L2YSA7 A0A0P8ZL89 W8B3F8 A0A293M3N6 A0A085LQ61 A0A0L7QJ97 A0A0L7QJ51 A0A131YM78 A0A139WA14 D7GYF3 A0A2G8JL30 D7EJX5 A0A3L8DMT5 A0A154PMS5 A0A1Y1NAW7 A0A182G9J3 A0A182H518 A0A151JBY5 D6WTN9 A0A0A9YNN7 A0A139W8G7 A0A293M349 A0A085N3N9 A0A182HB40 A0A2L2YTR5 A0A085NTM8 A0A2J7PD24 A0A151IIJ9 A0A026VX87 A0A0J7NAK0 A0A182H3L9 A0A154PT26 A0A0L8GMC8
Pubmed
EMBL
KL363297
KFD48322.1
KL367555
KFD64488.1
KL367519
KFD66949.1
+ More
LBMM01012479 KMQ86203.1 LBMM01013103 KMQ85816.1 KQ980726 KYN14011.1 KQ981018 KYN10257.1 LBMM01009142 KMQ88358.1 LBMM01012075 KMQ86422.1 NEVH01000707 PNF43124.1 GEZM01017686 JAV90494.1 NWSH01002598 PCG67884.1 KQ979048 KYN23218.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 KQ414987 KOC58988.1 NEVH01024424 PNF17276.1 NEVH01011195 PNF31836.1 LBMM01018089 KMQ83878.1 KQ981301 KYN43041.1 ACPB03037918 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 NEVH01013558 PNF28672.1 KK111495 EZA46390.1 MRZV01001127 MRZV01000236 PIK40395.1 PIK54731.1 NEVH01003759 PNF40011.1 GL447057 EFN86960.1 NEVH01014357 PNF27922.1 KQ434816 KZC06854.1 ODYU01011787 SOQ57731.1 NWSH01000267 PCG77880.1 KQ434784 KZC04993.1 NEVH01023981 PNF17578.1 MRZV01000025 PIK61772.1 GEZM01036621 JAV82564.1 GDQN01009305 JAT81749.1 NEVH01026435 PNF14140.1 GEDV01006140 JAP82417.1 LKEY01014428 KYN50445.1 KQ979024 KYN24351.1 NWSH01001575 PCG70822.1 AAGJ04150373 CH905996 KPU81830.1 KQ414874 KOC59935.1 IAAA01053498 IAAA01053499 LAA11042.1 CH902629 KPU75574.1 GAMC01010905 JAB95650.1 GFWV01009909 MAA34638.1 KL363340 KFD47107.1 KQ415240 KOC58703.1 LHQN01026442 KQ414593 KOC58672.1 KOC70162.1 GEDV01008183 JAP80374.1 KQ971745 KYB24749.1 GG695519 EFA13383.1 MRZV01001674 PIK36447.1 KQ971759 EFA12902.2 QOIP01000006 RLU21744.1 KQ434992 KZC13185.1 GEZM01010484 JAV94030.1 JXUM01049417 KQ561603 KXJ78027.1 JXUM01111110 KQ565471 KXJ70931.1 KQ979103 KYN22624.1 KQ971355 EFA07205.2 GBHO01008962 JAG34642.1 KQ973343 KXZ75568.1 GFWV01009910 MAA34639.1 KL367562 KFD64085.1 JXUM01031600 KQ560927 KXJ80313.1 IAAA01048245 LAA10725.1 KL367475 KFD72824.1 NEVH01026394 PNF14234.1 KQ977516 KYN02153.1 KK107653 EZA48265.1 LBMM01007548 KMQ89630.1 JXUM01107723 KQ565168 KXJ71261.1 KQ435204 KZC15052.1 KQ421206 KOF78131.1
LBMM01012479 KMQ86203.1 LBMM01013103 KMQ85816.1 KQ980726 KYN14011.1 KQ981018 KYN10257.1 LBMM01009142 KMQ88358.1 LBMM01012075 KMQ86422.1 NEVH01000707 PNF43124.1 GEZM01017686 JAV90494.1 NWSH01002598 PCG67884.1 KQ979048 KYN23218.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 KQ414987 KOC58988.1 NEVH01024424 PNF17276.1 NEVH01011195 PNF31836.1 LBMM01018089 KMQ83878.1 KQ981301 KYN43041.1 ACPB03037918 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 NEVH01013558 PNF28672.1 KK111495 EZA46390.1 MRZV01001127 MRZV01000236 PIK40395.1 PIK54731.1 NEVH01003759 PNF40011.1 GL447057 EFN86960.1 NEVH01014357 PNF27922.1 KQ434816 KZC06854.1 ODYU01011787 SOQ57731.1 NWSH01000267 PCG77880.1 KQ434784 KZC04993.1 NEVH01023981 PNF17578.1 MRZV01000025 PIK61772.1 GEZM01036621 JAV82564.1 GDQN01009305 JAT81749.1 NEVH01026435 PNF14140.1 GEDV01006140 JAP82417.1 LKEY01014428 KYN50445.1 KQ979024 KYN24351.1 NWSH01001575 PCG70822.1 AAGJ04150373 CH905996 KPU81830.1 KQ414874 KOC59935.1 IAAA01053498 IAAA01053499 LAA11042.1 CH902629 KPU75574.1 GAMC01010905 JAB95650.1 GFWV01009909 MAA34638.1 KL363340 KFD47107.1 KQ415240 KOC58703.1 LHQN01026442 KQ414593 KOC58672.1 KOC70162.1 GEDV01008183 JAP80374.1 KQ971745 KYB24749.1 GG695519 EFA13383.1 MRZV01001674 PIK36447.1 KQ971759 EFA12902.2 QOIP01000006 RLU21744.1 KQ434992 KZC13185.1 GEZM01010484 JAV94030.1 JXUM01049417 KQ561603 KXJ78027.1 JXUM01111110 KQ565471 KXJ70931.1 KQ979103 KYN22624.1 KQ971355 EFA07205.2 GBHO01008962 JAG34642.1 KQ973343 KXZ75568.1 GFWV01009910 MAA34639.1 KL367562 KFD64085.1 JXUM01031600 KQ560927 KXJ80313.1 IAAA01048245 LAA10725.1 KL367475 KFD72824.1 NEVH01026394 PNF14234.1 KQ977516 KYN02153.1 KK107653 EZA48265.1 LBMM01007548 KMQ89630.1 JXUM01107723 KQ565168 KXJ71261.1 KQ435204 KZC15052.1 KQ421206 KOF78131.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A085LTM6
A0A085N4U2
A0A085NBV3
A0A0J7K6V3
A0A0J7K6I6
A0A151IZ47
+ More
A0A151ITG2 A0A0J7KD10 A0A0J7K7T5 A0A2J7RQK2 A0A1Y1MXV9 A0A2A4J8M7 A0A151JCH0 A0A1W4XBP5 A0A0L7QJB5 A0A0L7QK36 A0A2J7PLR6 A0A2J7QTC8 A0A0J7K0A7 A0A151K0A8 T1IE13 A0A0J7K6V1 A0A2J7QJD3 A0A026VRX8 A0A2G8L3A3 A0A2J7RGP3 E2BBH0 A0A2J7QH56 A0A154P674 A0A2H1WXE7 A0A2A4K2H4 A0A154NZG0 A0A2J7PML8 A0A2G8LNJ3 A0A1Y1MA44 A0A1E1W488 A0A2J7PCS9 A0A1S3CY78 A0A131YTY1 A0A151K2U9 A0A151JF00 A0A2A4JHF4 W4YWZ1 A0A1I8Q981 A0A0N5DP72 A0A0P8Y533 A0A0L7QMU1 A0A2L2YSA7 A0A0P8ZL89 W8B3F8 A0A293M3N6 A0A085LQ61 A0A0L7QJ97 A0A0L7QJ51 A0A131YM78 A0A139WA14 D7GYF3 A0A2G8JL30 D7EJX5 A0A3L8DMT5 A0A154PMS5 A0A1Y1NAW7 A0A182G9J3 A0A182H518 A0A151JBY5 D6WTN9 A0A0A9YNN7 A0A139W8G7 A0A293M349 A0A085N3N9 A0A182HB40 A0A2L2YTR5 A0A085NTM8 A0A2J7PD24 A0A151IIJ9 A0A026VX87 A0A0J7NAK0 A0A182H3L9 A0A154PT26 A0A0L8GMC8
A0A151ITG2 A0A0J7KD10 A0A0J7K7T5 A0A2J7RQK2 A0A1Y1MXV9 A0A2A4J8M7 A0A151JCH0 A0A1W4XBP5 A0A0L7QJB5 A0A0L7QK36 A0A2J7PLR6 A0A2J7QTC8 A0A0J7K0A7 A0A151K0A8 T1IE13 A0A0J7K6V1 A0A2J7QJD3 A0A026VRX8 A0A2G8L3A3 A0A2J7RGP3 E2BBH0 A0A2J7QH56 A0A154P674 A0A2H1WXE7 A0A2A4K2H4 A0A154NZG0 A0A2J7PML8 A0A2G8LNJ3 A0A1Y1MA44 A0A1E1W488 A0A2J7PCS9 A0A1S3CY78 A0A131YTY1 A0A151K2U9 A0A151JF00 A0A2A4JHF4 W4YWZ1 A0A1I8Q981 A0A0N5DP72 A0A0P8Y533 A0A0L7QMU1 A0A2L2YSA7 A0A0P8ZL89 W8B3F8 A0A293M3N6 A0A085LQ61 A0A0L7QJ97 A0A0L7QJ51 A0A131YM78 A0A139WA14 D7GYF3 A0A2G8JL30 D7EJX5 A0A3L8DMT5 A0A154PMS5 A0A1Y1NAW7 A0A182G9J3 A0A182H518 A0A151JBY5 D6WTN9 A0A0A9YNN7 A0A139W8G7 A0A293M349 A0A085N3N9 A0A182HB40 A0A2L2YTR5 A0A085NTM8 A0A2J7PD24 A0A151IIJ9 A0A026VX87 A0A0J7NAK0 A0A182H3L9 A0A154PT26 A0A0L8GMC8
Ontologies
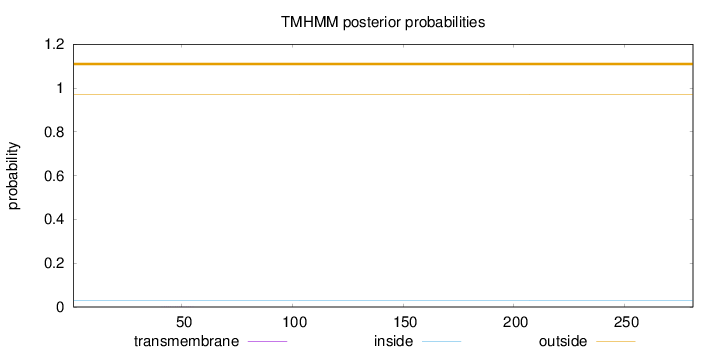
Topology
Length:
281
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00386
Exp number, first 60 AAs:
0.00326
Total prob of N-in:
0.02842
outside
1 - 281
Population Genetic Test Statistics
Pi
13.214659
Theta
24.646897
Tajima's D
-1.414493
CLR
7.136108
CSRT
0.0697465126743663
Interpretation
Uncertain
