Pre Gene Modal
BGIBMGA004208
Annotation
homocysteine_S-methyltransferase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.117 Nuclear Reliability : 1.249
Sequence
CDS
ATGAGACTAGCTATTTTTCGAACAGCGCTTCAATATTTAATAAGTCACACAGTTACGTATGCAACAGTGCTAAAAAGTATTAAAACTGCTTATGATAACGCTATAGAAATACGGGATGAAAGAATTTCACAGTTTATCGTACAGGATGTGTGTTGCAATAATTTACGTACTATAAACTTATTCTTTTATGAAATAATGTCGAATTTTGAAGTATCTTCAAAGACAGTCTTTGTGTTGGATGGAGGATTCTCAACACAGTTGACCTGTCACGCCGGTCATACTGCTGATGGCGATCCTCTTTGGAGTGCACGATTTTTAAAGACACACCCTCAAGATGTTATTAACACACATCTGGACTTCCTTCGTGCTGGCTCTGACATCATCGAAACAAATACTTACCAAGCCTCCGTAGATGGATTTGTTAAGCATCTGAACTTAACTGTCGAAGAGAGCTACGAACTGATAAAAAGTGCTGTTGAATTTGCAAGAACTGCACGCGACTTATACTTGCAAGAATGCCAAGAATCAAACCTTTCAGGTCGGAAACCACTCATAGCTGGTTCGGTAGGTCCTTATGGTGCATATTTACATGACACATCAGAATACACCGGTAATTATGCTGATAATACAACAAAGGAAACCATAAAAAATTGGCATAGGACAAGAATTCAAGCATTAGTAGAGGCAGGTGTAGATATATTGGCGTTCGAAACAATACCATGCCAAAAAGAAGCTGAAGCACTTGTAGAAATATTAAAAGAATATCCTAATATGAAAGCATGGTTATCATTTAGCTGCAAAAATGAGACTTCCTTAGCACATGGAGAAAATTTTCAAAATGTTGCCAAAAAATGTTGGAAATCAAACCCAGACCAGTTAATAGCCATCGGAGTCAATTGCTGCTCTCCGAAAATAGTGACTGAACTCTTTAAAGATATAAACAATGATCAAGAAACTTCCATACAATTTATCACATACCCTAACTCAGGAGAAACTTATGACCACAAACTTGGGTGGACTGAATCGGACAAATGTGAAAGCTTACATAATTTTGTTGCTGAATGGCTTGACCTTGGAGTTAGATACATAGACAGATCTTTGGTTGAAAAAAAACAGTCTATGAATAAAGTCTTCGGAATATCTGTCAAAAAGAAACAACGGAATGATCAAATTCAAAACTTGCTCAAAGAAAATAATGATCTCAAAGAAAAAAACAATAAATTAGTATTAGAAATGGATAAATTAAAAAAAGAATTTGGAAAGATGAAAGATGACAAATTCTCCGACTACCTCTCTTTGATGAAAGAGCGAGATGCAAGATACGCTTTATATTTCGAAAACCTAACTCTTCAAACGAAGCTTAAAGACCTTGATGGTTCATTTTGCCCGTCCAATGATGCATATGGAGATCCTGTGGTGCTGAGGATAGCCCTGGATAGATGCAGGTGA
Protein
MRLAIFRTALQYLISHTVTYATVLKSIKTAYDNAIEIRDERISQFIVQDVCCNNLRTINLFFYEIMSNFEVSSKTVFVLDGGFSTQLTCHAGHTADGDPLWSARFLKTHPQDVINTHLDFLRAGSDIIETNTYQASVDGFVKHLNLTVEESYELIKSAVEFARTARDLYLQECQESNLSGRKPLIAGSVGPYGAYLHDTSEYTGNYADNTTKETIKNWHRTRIQALVEAGVDILAFETIPCQKEAEALVEILKEYPNMKAWLSFSCKNETSLAHGENFQNVAKKCWKSNPDQLIAIGVNCCSPKIVTELFKDINNDQETSIQFITYPNSGETYDHKLGWTESDKCESLHNFVAEWLDLGVRYIDRSLVEKKQSMNKVFGISVKKKQRNDQIQNLLKENNDLKEKNNKLVLEMDKLKKEFGKMKDDKFSDYLSLMKERDARYALYFENLTLQTKLKDLDGSFCPSNDAYGDPVVLRIALDRCR
Summary
Cofactor
Zn(2+)
EMBL
Proteomes
Pfam
PF02574 S-methyl_trans
SUPFAM
SSF82282
SSF82282
Gene 3D
ProteinModelPortal
PDB
5DMN
E-value=5.40172e-47,
Score=474
Ontologies
PATHWAY
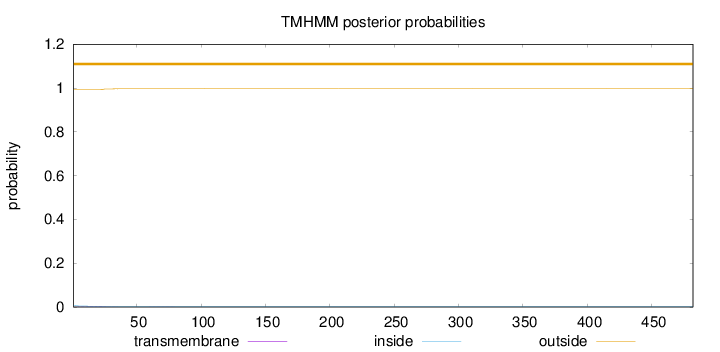
Topology
Length:
482
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07333
Exp number, first 60 AAs:
0.06959
Total prob of N-in:
0.00635
outside
1 - 482
Population Genetic Test Statistics
Pi
6.029627
Theta
14.011545
Tajima's D
-1.039099
CLR
0.42774
CSRT
0.130493475326234
Interpretation
Uncertain
