Gene
KWMTBOMO11752
Pre Gene Modal
BGIBMGA004156
Annotation
hypothetical_protein_KGM_06377_[Danaus_plexippus]
Full name
Non-specific serine/threonine protein kinase
Location in the cell
Mitochondrial Reliability : 3.005
Sequence
CDS
ATGAGAGCCATGACAAAGAGTAAGAATCAAGTGTACAGTGTTGGGAACTATCTGATGACCGGCAGAGTTTTGGGGAAGGGACACTTCGCACGCGTCGAAGAAGCCACACACAGAATTATAGGCGCTAAGGTGGCAGTTAAAGTGATAGACCTAACATGTATAAAAGAGGAATACGCACGCAGGAACCTGCATCGCGAACCAAGAGTGATGGCGAGACTGCGGCATCCTTGCATCGCTGCTCTGTACGAAACGATGATGCACGGGCCGCGCCTTTACGTGGTGATGGAGGCGGCGGGCGGCGGCGACCTGTGCTCGCACGTGCTGGGGGCTCGCGGGGGCGCGCGGGGCCTCCCCGAGCACAGGGCACGTGCTCTCGCCGCACAGCTCGTCTCCGCCGTTCGGCACATGCATGCGCGTGCCGTGGTGCATCGGGATCTTAAAATGGAAAACATCATGTTGGACAGTACAAAGCAGTTTATAAAAATAGTCGATTTCGGGTTATCGAATCTTTGGAGCTCGAGCGGCGGGCTGAGGACGCCCTGCGGCTCCCTGGAGTACGCCGCGCCGGAACTCTTCATCGACGGAAGGAAATACGGGCCGGAGGTCGACCTCTGGAGCATGTGA
Protein
MRAMTKSKNQVYSVGNYLMTGRVLGKGHFARVEEATHRIIGAKVAVKVIDLTCIKEEYARRNLHREPRVMARLRHPCIAALYETMMHGPRLYVVMEAAGGGDLCSHVLGARGGARGLPEHRARALAAQLVSAVRHMHARAVVHRDLKMENIMLDSTKQFIKIVDFGLSNLWSSSGGLRTPCGSLEYAAPELFIDGRKYGPEVDLWSM
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the protein kinase superfamily.
Belongs to the peptidase C14A family.
Belongs to the peptidase C14A family.
Uniprot
A0A2H1W7M2
A0A212F3W9
A0A194QXY6
A0A194PP49
A0A3S2LAA9
H9J3R9
+ More
A0A2W1BIS9 A0A1W4WU57 A0A1W4WJB8 A0A1W4WJD5 A0A1W4WVE5 A0A1W4WV70 A0A139WEU1 A0A0L7LIU7 A0A139WF40 T1J935 A0A210QK57 V4B4G9 A0A2G8JPM3 A0A1Y1K8W7 A0A1X7VLU9 A0A1S3IJZ4 T1FXU7 A0A1Y1K9Q9 A0A2C9JHR6 A0A2C9JHR5 A0A2C9JHR7 A0A2C9JHT4 A0A2C9JHS3 A0A2C9JHR2 A0A1B6F977 K7IR62 A0A3L8DAT1 A0A158NIB8 R7TX05 A0A3M6T7Q7 T1G334 A0A195CNT2 A0A087VES0 A0A1S3IL01 A0A2T7NQP1 A0A093F3I0 A0A151JXT9 A0A1S8W1Z4 A0A093FC57 A0A091F1V5 A0A091H5K0 A0A1J1IJC2 A0A091SH21 A0A091TBV9 A0A0S7FV21 A0A091PQ03 A0A091I3R6 A0A093PNJ0 A0A091N1M2 A0A091NDB9 A0A091W104 A0A093HEW3 A0A0A0AP48 A0A093QJV4 A0A1A8N4B4 A0A093IBF0 A0A087R8L1 A0A093BZG1 A0A3P4R9Y3 F7G3W8 K7G3A7 A0A091LGG3 A0A1A8BJ32 F4NZ62 A0A093G5U0 A0A139AJS0 A0A1A7YFF5 A0A091F6F3 A0A0N4XKN9 A0A093PMY1 A0A1A8M1X1 A0A1A8L9I7 A0A3P7XSU3 A0A1A7Y7M9 A0A183FMV9 A0A091QL49 A0A0K0CVJ6 A0A0R3WK39 G1NLS8 U3JJG0 H3BAS6 A0A0D2VKD6 W5NHE2 A0A0N4VSA4 A0A2H2J0A5 R7UFG2 H0ZQH9 A0A151P543 A0A0N4XHD0 W5NHD8 Q8MVX4 A0A099ZH75 A0A087YNK5 A0A3B5LCQ5
A0A2W1BIS9 A0A1W4WU57 A0A1W4WJB8 A0A1W4WJD5 A0A1W4WVE5 A0A1W4WV70 A0A139WEU1 A0A0L7LIU7 A0A139WF40 T1J935 A0A210QK57 V4B4G9 A0A2G8JPM3 A0A1Y1K8W7 A0A1X7VLU9 A0A1S3IJZ4 T1FXU7 A0A1Y1K9Q9 A0A2C9JHR6 A0A2C9JHR5 A0A2C9JHR7 A0A2C9JHT4 A0A2C9JHS3 A0A2C9JHR2 A0A1B6F977 K7IR62 A0A3L8DAT1 A0A158NIB8 R7TX05 A0A3M6T7Q7 T1G334 A0A195CNT2 A0A087VES0 A0A1S3IL01 A0A2T7NQP1 A0A093F3I0 A0A151JXT9 A0A1S8W1Z4 A0A093FC57 A0A091F1V5 A0A091H5K0 A0A1J1IJC2 A0A091SH21 A0A091TBV9 A0A0S7FV21 A0A091PQ03 A0A091I3R6 A0A093PNJ0 A0A091N1M2 A0A091NDB9 A0A091W104 A0A093HEW3 A0A0A0AP48 A0A093QJV4 A0A1A8N4B4 A0A093IBF0 A0A087R8L1 A0A093BZG1 A0A3P4R9Y3 F7G3W8 K7G3A7 A0A091LGG3 A0A1A8BJ32 F4NZ62 A0A093G5U0 A0A139AJS0 A0A1A7YFF5 A0A091F6F3 A0A0N4XKN9 A0A093PMY1 A0A1A8M1X1 A0A1A8L9I7 A0A3P7XSU3 A0A1A7Y7M9 A0A183FMV9 A0A091QL49 A0A0K0CVJ6 A0A0R3WK39 G1NLS8 U3JJG0 H3BAS6 A0A0D2VKD6 W5NHE2 A0A0N4VSA4 A0A2H2J0A5 R7UFG2 H0ZQH9 A0A151P543 A0A0N4XHD0 W5NHD8 Q8MVX4 A0A099ZH75 A0A087YNK5 A0A3B5LCQ5
EC Number
2.7.11.1
Pubmed
EMBL
ODYU01006851
SOQ49070.1
AGBW02010485
OWR48429.1
KQ460949
KPJ10403.1
+ More
KQ459598 KPI94524.1 RSAL01000070 RVE49197.1 BABH01028870 BABH01028871 KZ150291 PZC71583.1 KQ971354 KYB26480.1 JTDY01001014 KOB75126.1 KYB26479.1 JH431969 NEDP02003233 OWF49134.1 KB199650 ESP05363.1 MRZV01001462 PIK37722.1 GEZM01091323 JAV56911.1 AMQM01000580 KB096324 ESO06195.1 GEZM01091324 JAV56910.1 GECZ01022971 JAS46798.1 QOIP01000011 RLU17243.1 ADTU01016343 ADTU01016344 ADTU01016345 ADTU01016346 ADTU01016347 AMQN01002100 AMQN01002101 AMQN01002102 KB308242 ELT98137.1 RCHS01004154 RMX37318.1 AMQM01003892 KB096365 ESO05214.1 KQ977481 KYN02398.1 KL490572 KFO11112.1 PZQS01000010 PVD23463.1 KK381644 KFV48756.1 KQ981578 KYN39745.1 LYON01000243 OON08524.1 KK627808 KFV54135.1 KK719548 KFO64098.1 KL522405 KFO90067.1 CVRI01000049 CRK99158.1 KK819579 KFQ39555.1 KK444851 KFQ71322.1 GBYX01461288 JAO20280.1 KK663437 KFQ09675.1 KL218085 KFP02065.1 KL670077 KFW78011.1 KL374453 KFP82774.1 KK845318 KFP87072.1 KL411541 KFR09347.1 KL206020 KFV77965.1 KL872360 KGL95323.1 KL421770 KFW89163.1 HAEG01000425 SBR63629.1 KK553951 KFV96906.1 KL226213 KFM09815.1 KL451785 KFV06224.1 CYRY02039461 VCX30861.1 AGCU01154743 AGCU01154744 AGCU01154745 KK751705 KFP41996.1 HADZ01002695 SBP66636.1 GL882881 EGF81831.1 KL214891 KFV62139.1 KQ965749 KXS17056.1 HADX01006652 SBP28884.1 KFO64099.1 UYSL01004217 VDL66681.1 KL225531 KFW73532.1 HAEF01010522 SBR50541.1 HAEF01003162 SBR40544.1 UZAH01026246 VDO77706.1 HADX01003733 SBP25965.1 KK702612 KFQ28280.1 UYWX01000171 VDM17410.1 AGTO01010785 AFYH01061772 AFYH01061773 AFYH01061774 KE346361 KJE90477.1 AHAT01015758 UZAF01000091 VDO04657.1 AMQN01008794 KB303969 ELU02518.1 ABQF01000737 AKHW03000903 KYO44090.1 UYSL01001865 VDL65522.1 AF457198 AAM73857.1 KL892817 KGL80165.1 AYCK01020175 AYCK01020176 AYCK01020177
KQ459598 KPI94524.1 RSAL01000070 RVE49197.1 BABH01028870 BABH01028871 KZ150291 PZC71583.1 KQ971354 KYB26480.1 JTDY01001014 KOB75126.1 KYB26479.1 JH431969 NEDP02003233 OWF49134.1 KB199650 ESP05363.1 MRZV01001462 PIK37722.1 GEZM01091323 JAV56911.1 AMQM01000580 KB096324 ESO06195.1 GEZM01091324 JAV56910.1 GECZ01022971 JAS46798.1 QOIP01000011 RLU17243.1 ADTU01016343 ADTU01016344 ADTU01016345 ADTU01016346 ADTU01016347 AMQN01002100 AMQN01002101 AMQN01002102 KB308242 ELT98137.1 RCHS01004154 RMX37318.1 AMQM01003892 KB096365 ESO05214.1 KQ977481 KYN02398.1 KL490572 KFO11112.1 PZQS01000010 PVD23463.1 KK381644 KFV48756.1 KQ981578 KYN39745.1 LYON01000243 OON08524.1 KK627808 KFV54135.1 KK719548 KFO64098.1 KL522405 KFO90067.1 CVRI01000049 CRK99158.1 KK819579 KFQ39555.1 KK444851 KFQ71322.1 GBYX01461288 JAO20280.1 KK663437 KFQ09675.1 KL218085 KFP02065.1 KL670077 KFW78011.1 KL374453 KFP82774.1 KK845318 KFP87072.1 KL411541 KFR09347.1 KL206020 KFV77965.1 KL872360 KGL95323.1 KL421770 KFW89163.1 HAEG01000425 SBR63629.1 KK553951 KFV96906.1 KL226213 KFM09815.1 KL451785 KFV06224.1 CYRY02039461 VCX30861.1 AGCU01154743 AGCU01154744 AGCU01154745 KK751705 KFP41996.1 HADZ01002695 SBP66636.1 GL882881 EGF81831.1 KL214891 KFV62139.1 KQ965749 KXS17056.1 HADX01006652 SBP28884.1 KFO64099.1 UYSL01004217 VDL66681.1 KL225531 KFW73532.1 HAEF01010522 SBR50541.1 HAEF01003162 SBR40544.1 UZAH01026246 VDO77706.1 HADX01003733 SBP25965.1 KK702612 KFQ28280.1 UYWX01000171 VDM17410.1 AGTO01010785 AFYH01061772 AFYH01061773 AFYH01061774 KE346361 KJE90477.1 AHAT01015758 UZAF01000091 VDO04657.1 AMQN01008794 KB303969 ELU02518.1 ABQF01000737 AKHW03000903 KYO44090.1 UYSL01001865 VDL65522.1 AF457198 AAM73857.1 KL892817 KGL80165.1 AYCK01020175 AYCK01020176 AYCK01020177
Proteomes
UP000007151
UP000053240
UP000053268
UP000283053
UP000005204
UP000192223
+ More
UP000007266 UP000037510 UP000242188 UP000030746 UP000230750 UP000007879 UP000085678 UP000015101 UP000076420 UP000002358 UP000279307 UP000005205 UP000014760 UP000275408 UP000078542 UP000245119 UP000078541 UP000189906 UP000052976 UP000183832 UP000054308 UP000053258 UP000053283 UP000053584 UP000053858 UP000053286 UP000002279 UP000007267 UP000007241 UP000053875 UP000070544 UP000038043 UP000271162 UP000054081 UP000050761 UP000035642 UP000046396 UP000274429 UP000001645 UP000016665 UP000008672 UP000008743 UP000018468 UP000038042 UP000268014 UP000005237 UP000007754 UP000050525 UP000053641 UP000028760 UP000261380
UP000007266 UP000037510 UP000242188 UP000030746 UP000230750 UP000007879 UP000085678 UP000015101 UP000076420 UP000002358 UP000279307 UP000005205 UP000014760 UP000275408 UP000078542 UP000245119 UP000078541 UP000189906 UP000052976 UP000183832 UP000054308 UP000053258 UP000053283 UP000053584 UP000053858 UP000053286 UP000002279 UP000007267 UP000007241 UP000053875 UP000070544 UP000038043 UP000271162 UP000054081 UP000050761 UP000035642 UP000046396 UP000274429 UP000001645 UP000016665 UP000008672 UP000008743 UP000018468 UP000038042 UP000268014 UP000005237 UP000007754 UP000050525 UP000053641 UP000028760 UP000261380
PRIDE
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR036899 Ribosomal_L13_sf
IPR005822 Ribosomal_L13
IPR016129 Caspase_his_AS
IPR015917 Pept_C14A
IPR001309 Pept_C14_p20
IPR029030 Caspase-like_dom_sf
IPR002398 Pept_C14
IPR002138 Pept_C14_p10
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001772 KA1_dom
IPR028375 KA1/Ssp2_C
IPR015940 UBA
IPR009060 UBA-like_sf
IPR034671 Hunk
IPR033628 MARK3
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR036899 Ribosomal_L13_sf
IPR005822 Ribosomal_L13
IPR016129 Caspase_his_AS
IPR015917 Pept_C14A
IPR001309 Pept_C14_p20
IPR029030 Caspase-like_dom_sf
IPR002398 Pept_C14
IPR002138 Pept_C14_p10
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001772 KA1_dom
IPR028375 KA1/Ssp2_C
IPR015940 UBA
IPR009060 UBA-like_sf
IPR034671 Hunk
IPR033628 MARK3
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1W7M2
A0A212F3W9
A0A194QXY6
A0A194PP49
A0A3S2LAA9
H9J3R9
+ More
A0A2W1BIS9 A0A1W4WU57 A0A1W4WJB8 A0A1W4WJD5 A0A1W4WVE5 A0A1W4WV70 A0A139WEU1 A0A0L7LIU7 A0A139WF40 T1J935 A0A210QK57 V4B4G9 A0A2G8JPM3 A0A1Y1K8W7 A0A1X7VLU9 A0A1S3IJZ4 T1FXU7 A0A1Y1K9Q9 A0A2C9JHR6 A0A2C9JHR5 A0A2C9JHR7 A0A2C9JHT4 A0A2C9JHS3 A0A2C9JHR2 A0A1B6F977 K7IR62 A0A3L8DAT1 A0A158NIB8 R7TX05 A0A3M6T7Q7 T1G334 A0A195CNT2 A0A087VES0 A0A1S3IL01 A0A2T7NQP1 A0A093F3I0 A0A151JXT9 A0A1S8W1Z4 A0A093FC57 A0A091F1V5 A0A091H5K0 A0A1J1IJC2 A0A091SH21 A0A091TBV9 A0A0S7FV21 A0A091PQ03 A0A091I3R6 A0A093PNJ0 A0A091N1M2 A0A091NDB9 A0A091W104 A0A093HEW3 A0A0A0AP48 A0A093QJV4 A0A1A8N4B4 A0A093IBF0 A0A087R8L1 A0A093BZG1 A0A3P4R9Y3 F7G3W8 K7G3A7 A0A091LGG3 A0A1A8BJ32 F4NZ62 A0A093G5U0 A0A139AJS0 A0A1A7YFF5 A0A091F6F3 A0A0N4XKN9 A0A093PMY1 A0A1A8M1X1 A0A1A8L9I7 A0A3P7XSU3 A0A1A7Y7M9 A0A183FMV9 A0A091QL49 A0A0K0CVJ6 A0A0R3WK39 G1NLS8 U3JJG0 H3BAS6 A0A0D2VKD6 W5NHE2 A0A0N4VSA4 A0A2H2J0A5 R7UFG2 H0ZQH9 A0A151P543 A0A0N4XHD0 W5NHD8 Q8MVX4 A0A099ZH75 A0A087YNK5 A0A3B5LCQ5
A0A2W1BIS9 A0A1W4WU57 A0A1W4WJB8 A0A1W4WJD5 A0A1W4WVE5 A0A1W4WV70 A0A139WEU1 A0A0L7LIU7 A0A139WF40 T1J935 A0A210QK57 V4B4G9 A0A2G8JPM3 A0A1Y1K8W7 A0A1X7VLU9 A0A1S3IJZ4 T1FXU7 A0A1Y1K9Q9 A0A2C9JHR6 A0A2C9JHR5 A0A2C9JHR7 A0A2C9JHT4 A0A2C9JHS3 A0A2C9JHR2 A0A1B6F977 K7IR62 A0A3L8DAT1 A0A158NIB8 R7TX05 A0A3M6T7Q7 T1G334 A0A195CNT2 A0A087VES0 A0A1S3IL01 A0A2T7NQP1 A0A093F3I0 A0A151JXT9 A0A1S8W1Z4 A0A093FC57 A0A091F1V5 A0A091H5K0 A0A1J1IJC2 A0A091SH21 A0A091TBV9 A0A0S7FV21 A0A091PQ03 A0A091I3R6 A0A093PNJ0 A0A091N1M2 A0A091NDB9 A0A091W104 A0A093HEW3 A0A0A0AP48 A0A093QJV4 A0A1A8N4B4 A0A093IBF0 A0A087R8L1 A0A093BZG1 A0A3P4R9Y3 F7G3W8 K7G3A7 A0A091LGG3 A0A1A8BJ32 F4NZ62 A0A093G5U0 A0A139AJS0 A0A1A7YFF5 A0A091F6F3 A0A0N4XKN9 A0A093PMY1 A0A1A8M1X1 A0A1A8L9I7 A0A3P7XSU3 A0A1A7Y7M9 A0A183FMV9 A0A091QL49 A0A0K0CVJ6 A0A0R3WK39 G1NLS8 U3JJG0 H3BAS6 A0A0D2VKD6 W5NHE2 A0A0N4VSA4 A0A2H2J0A5 R7UFG2 H0ZQH9 A0A151P543 A0A0N4XHD0 W5NHD8 Q8MVX4 A0A099ZH75 A0A087YNK5 A0A3B5LCQ5
PDB
2HAK
E-value=3.32538e-34,
Score=359
Ontologies
GO
PANTHER
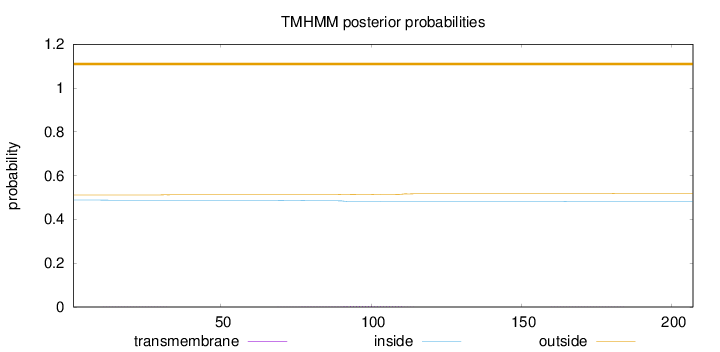
Topology
Length:
207
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13647
Exp number, first 60 AAs:
0.02014
Total prob of N-in:
0.48843
outside
1 - 207
Population Genetic Test Statistics
Pi
21.760544
Theta
17.753198
Tajima's D
0.859354
CLR
0.559813
CSRT
0.622618869056547
Interpretation
Uncertain
