Gene
KWMTBOMO11748 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004157
Annotation
1607133A_egg_specific_protein
Full name
Lipase
Location in the cell
Nuclear Reliability : 3.307
Sequence
CDS
ATGAAGACTATATACGCATTGCTGTGTCTGACGCTGGTGCAGAGCATCTCCTGCAGCATCTTCATGACGAAACAACACAGTCAGGATGACATCATTCAACACCCTCTGGACTATGTCGAACAGCAGATCCATCAGCAGAAACAAAAACTACACAAGCAAACCTTGAACAAGAGGAGCCACCAGCACTCTGATTCGGATTCGGATTCGGCGTCTCGTGCGGCGGCGTCACATTCAGCCTCCCAATCGTCGAGTTCACAAAGCTCCAGTTCACAAGAGGATGAAACTAAGCAGGTGCACGATAAGATGAACGTGAAACACCACTCGCCGGTGTATTCTGTCATTATGAAACTCAAGAAAGAAGTTGATATCAATCACGGCGATTCCGTCGTTTGGAAGAATATAGAAATGGCCTCCGGCCCTAACTCGCCGGTTCAGACAGAGCAAGATATTGAGGATATTTTCGGTGACTCCCTAAAGACGTGGGATCATTTCACTGACGATGCAAAGAAAAATACCTTCCACGACGCTATCAGTGAAACTCAAAGGGAAAACAATGAGGACTTCCACCTAAACGCTACTGAGCTGCTCAAGAAACATCAATACCCCGTAGAAGAACACACGGTCGCCACCGACGACGGCTACCATTTGACTGTCCTGCGCATTCCACCCACGCACCAAACCAGGGACGATAAGAAGAAGCCGGTCGCGCTTCTAATGCACGGCTTACTCGGAAGCGCTGACGACTGGTTACTGATGGGTCCCAGTAAGTCACTCGCTTACATGCTCTGTGACGCCGGCTACGACGTATGGCTGGGTAATGTTCGTGGAAACAAATATTCCCGCTCTCACGTCAGCAAGCACCCAGCACTCAATGACTTCTGGAAGTTTAGCAATGACGAGATCGCTCTTCACGACTTACCCGCTATAATTGACCACGTTTTGGATATTAGCGGCCAAGAGAGACTTCATTACATAGGCCATTCTCAAGGCGCGACCACCTTCTTCGCCCTGATGTCCGAACAGCCTTCGTACAACGAAAAGATCGTTTCGATGCACGCGTTGTCTCCTATTGTTTACATGAATTATGTACGCTCGCCCCTCTTCCGTATGATCGCGCCCACGAGCAAGTTCTACCAGTATATACACGACCAAGTCGGTCACGGAGCCTTCGAACCTGGCAAACACCTGATCGAAACCTTCGGCGGCGCCGCGTGCAGGGAAAAACTCGGTTGCAGACACGTCTGTAACAACTTGAACTACGTCATATCGGGTATCAACGTTTACAACCAGGATGCGGATATAGTTCCCGTTGTGATGGCCCACCTGCCAGCCGGCACATCCGCCCGGGTCATGAAACAATACGGTCAGAATGTGGCGTCGCACGATTTTAGAAAATACAACTACGGAGCAGAAACCAACATGAAAGTGTACGGCACTTCGGAACCACCTAGTTACGACTTGAGCAAAGTCAGCGCGCCTGTCAATCTTTACCACAGCCACGATGCCTGGTTGGCCCATCCCAAGGACGTGGAGAAACTCCAAGAAAACCTGCCTAATGTGAAGCAGTCTTTCGAAGTTCCAGAGCAACAACACTTCACGGACCTGGACTTCCAATTCTCGAAGAAAGCCCCCGATACCGTATACCAGAAACTGATGGAAAACATGCAGAATAACTCATAA
Protein
MKTIYALLCLTLVQSISCSIFMTKQHSQDDIIQHPLDYVEQQIHQQKQKLHKQTLNKRSHQHSDSDSDSASRAAASHSASQSSSSQSSSSQEDETKQVHDKMNVKHHSPVYSVIMKLKKEVDINHGDSVVWKNIEMASGPNSPVQTEQDIEDIFGDSLKTWDHFTDDAKKNTFHDAISETQRENNEDFHLNATELLKKHQYPVEEHTVATDDGYHLTVLRIPPTHQTRDDKKKPVALLMHGLLGSADDWLLMGPSKSLAYMLCDAGYDVWLGNVRGNKYSRSHVSKHPALNDFWKFSNDEIALHDLPAIIDHVLDISGQERLHYIGHSQGATTFFALMSEQPSYNEKIVSMHALSPIVYMNYVRSPLFRMIAPTSKFYQYIHDQVGHGAFEPGKHLIETFGGAACREKLGCRHVCNNLNYVISGINVYNQDADIVPVVMAHLPAGTSARVMKQYGQNVASHDFRKYNYGAETNMKVYGTSEPPSYDLSKVSAPVNLYHSHDAWLAHPKDVEKLQENLPNVKQSFEVPEQQHFTDLDFQFSKKAPDTVYQKLMENMQNNS
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
EMBL
BABH01028866
D12521
BAA02091.1
NWSH01000119
PCG79375.1
KZ150162
+ More
PZC72687.1 JTDY01001496 KOB73732.1 ODYU01004598 SOQ44646.1 KQ459598 KPI94528.1 KQ460949 KPJ10407.1 MF067302 ASM61899.1 FJ744151 ACT53735.1 AF063014 AAC62229.1 AB081090 BAC66969.1 U69881 AAB09081.1 KQ460855 KPJ11879.1 KZ149971 PZC76037.1 KQ461190 KPJ07151.1 KPJ07150.1 GEZM01086956 JAV58943.1 AGBW02009374 OWR50972.1 GANO01002781 JAB57090.1 AGBW02008724 OWR52685.1
PZC72687.1 JTDY01001496 KOB73732.1 ODYU01004598 SOQ44646.1 KQ459598 KPI94528.1 KQ460949 KPJ10407.1 MF067302 ASM61899.1 FJ744151 ACT53735.1 AF063014 AAC62229.1 AB081090 BAC66969.1 U69881 AAB09081.1 KQ460855 KPJ11879.1 KZ149971 PZC76037.1 KQ461190 KPJ07151.1 KPJ07150.1 GEZM01086956 JAV58943.1 AGBW02009374 OWR50972.1 GANO01002781 JAB57090.1 AGBW02008724 OWR52685.1
Proteomes
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
PDB
1K8Q
E-value=5.53725e-63,
Score=613
Ontologies
PATHWAY
GO
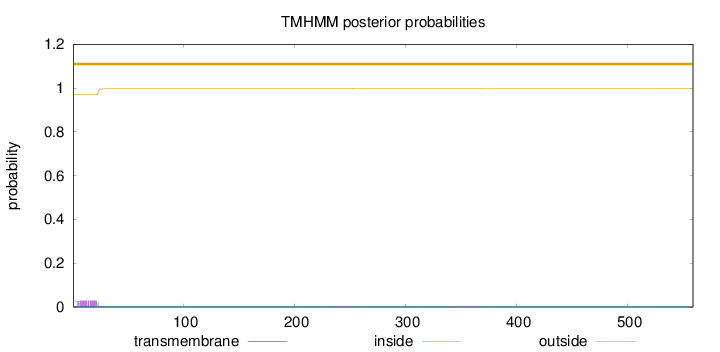
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.971661
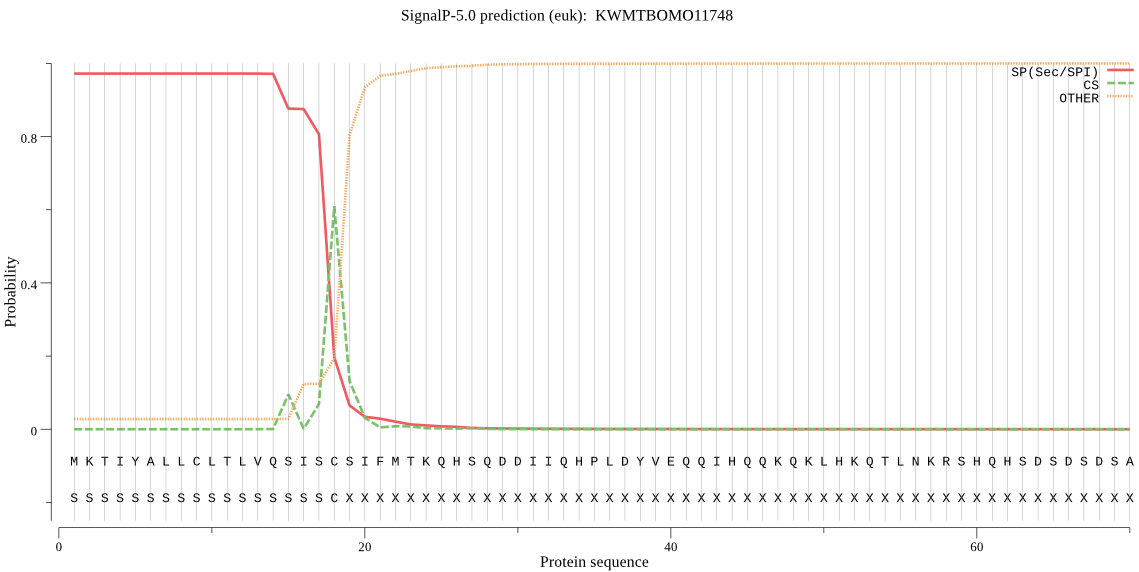
Length:
559
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.590139999999999
Exp number, first 60 AAs:
0.56259
Total prob of N-in:
0.02869
outside
1 - 559
Population Genetic Test Statistics
Pi
15.795923
Theta
13.268648
Tajima's D
0.45648
CLR
3.51098
CSRT
0.514424278786061
Interpretation
Uncertain
