Pre Gene Modal
BGIBMGA004158
Annotation
achintya_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.767
Sequence
CDS
ATGTTACCTCCCACGGCCGGTCTCTCACAAGAACAGCGCGACCGGACCGAAATGGCATTCCAACACCGACGTGACGTGAGGGAATTGACCCGAGAAATCATCAGGGACGCTCGCGTCGCTAACAGACAACAGTCCTCATCTCCTGGAGTAAGAAACCGTTTCCCTTCGACTTCATCGACGGACGAAAATGAGAGCGGCACAGACGGCGAGGCGGACCGGTCGCGCCGGCAGACCATCCTCCACGGAGTCTCCGGCATGCAGCCCCAGGGTCAGATAGTGACAATACGGAAACGTCGAGGGAACCTCGCCAAGCATTCGGTGAAGATATTGAAGCGATGGCTGTACGACCACAGGTACAACGCGTACCCGAGCGACGCCGAGAAAATAGCGCTCAGCCAGGAGGCCAATTTGAGTGTCTTGCAGGTATGTAACTGGTTTATAAACGCGAGACGTCGCATCTTGCCGGAGATGATACGTCGCGAGGGTCATGATCCGTTACACTACACGATCTCACGCCGCGGACGGAAACAACCCTCGGGCGCTTCCTCCAGCGCGTCCAACTCTGGAGTCGTTGTCGCTTCTTGGGACGGAGCTGATGTTGAGAATGATCCAGACCGCAGCGCAGAGCAAGAGTACACTGATGGTCTACTGGTGTATCGTAGTGAAGGAGATGACATTGCAGACGGTGAAGAAGGCTACTCCTCCAGTGCGGTCAGCGAGGAAGAAGTGAAATACGATCCTTCTGTGTGGCAGTCGGTCATCAGATACGGACCAGAAGATAAGGAGATACATCCCGCTGTCAGAGGTACGGCGTCGGCGGTGGTGACGGTGCGCACGGCGGAGGGGGCCGTGGAGGGCGACGGAGAGGGGGGTGAAGGGGAGGGAGAAGGCGGCACGCCGGGGGGCTGGCACCCACAACTCGCACATCTTCCGCAACTGCCGCACCCGCTCAGGAAACGGCTGCAGGTGGTCCCGGCGGCGGGCGAAGGCGAGCGCGACAAGTTCAAGTGCCTGTACCTGCTGGTGGAGACCGCCGTGGCCGTGCGCCAGCGCGAGAAGGAAGCCGACGAGCTGCCCGTCTAG
Protein
MLPPTAGLSQEQRDRTEMAFQHRRDVRELTREIIRDARVANRQQSSSPGVRNRFPSTSSTDENESGTDGEADRSRRQTILHGVSGMQPQGQIVTIRKRRGNLAKHSVKILKRWLYDHRYNAYPSDAEKIALSQEANLSVLQVCNWFINARRRILPEMIRREGHDPLHYTISRRGRKQPSGASSSASNSGVVVASWDGADVENDPDRSAEQEYTDGLLVYRSEGDDIADGEEGYSSSAVSEEEVKYDPSVWQSVIRYGPEDKEIHPAVRGTASAVVTVRTAEGAVEGDGEGGEGEGEGGTPGGWHPQLAHLPQLPHPLRKRLQVVPAAGEGERDKFKCLYLLVETAVAVRQREKEADELPV
Summary
Similarity
Belongs to the adenylate kinase family.
Belongs to the MCM family.
Belongs to the MCM family.
Uniprot
C0KWE9
H9J3S1
B8Y9D1
A0A2H1VUX8
A0A2A4K5S1
A0A212F3W8
+ More
A0A3S2NUR2 A0A194PP62 A0A1E1WVG8 A0A2W1BIY9 A0A194QXZ1 A0A2A4KAY0 A0A2H1VQ46 A0A2W1BHR6 A0A0L7LDY1 H9J9C4 A0A0T6B0Q6 A0A1W4WK63 A0A1W4WW72 D6WTB7 A0A1Y1L0S3 U4UBP6 N6U0C7 J3JYU1 A0A067RN17 A0A0C9R0G7 A0A2A3E924 A0A2J7R5E8 A0A0R1DUE7 Q7K380 Q7JR08
A0A3S2NUR2 A0A194PP62 A0A1E1WVG8 A0A2W1BIY9 A0A194QXZ1 A0A2A4KAY0 A0A2H1VQ46 A0A2W1BHR6 A0A0L7LDY1 H9J9C4 A0A0T6B0Q6 A0A1W4WK63 A0A1W4WW72 D6WTB7 A0A1Y1L0S3 U4UBP6 N6U0C7 J3JYU1 A0A067RN17 A0A0C9R0G7 A0A2A3E924 A0A2J7R5E8 A0A0R1DUE7 Q7K380 Q7JR08
Pubmed
EMBL
FJ643616
ACN38265.1
BABH01028865
FJ502823
FJ502824
ACL54971.1
+ More
ODYU01004598 SOQ44647.1 NWSH01000119 PCG79376.1 AGBW02010485 OWR48424.1 RSAL01000070 RVE49194.1 KQ459598 KPI94529.1 GDQN01000066 JAT90988.1 KZ150162 PZC72700.1 KQ460949 KPJ10408.1 NWSH01000011 PCG80900.1 ODYU01003780 SOQ42965.1 KZ150052 PZC74388.1 JTDY01001496 KOB73733.1 BABH01004466 LJIG01016339 KRT80898.1 KQ971352 EFA06692.1 GEZM01068010 JAV67263.1 KB632281 ERL91329.1 APGK01054085 KB741247 ENN71992.1 BT128422 AEE63379.1 KK852527 KDR22010.1 GBYB01000296 JAG70063.1 KZ288322 PBC28225.1 NEVH01006994 PNF36065.1 CM000158 KRJ99442.1 AE013599 AY058750 AAG22280.2 AAL13979.1 BT003510 AAF58497.2 AAO39514.1
ODYU01004598 SOQ44647.1 NWSH01000119 PCG79376.1 AGBW02010485 OWR48424.1 RSAL01000070 RVE49194.1 KQ459598 KPI94529.1 GDQN01000066 JAT90988.1 KZ150162 PZC72700.1 KQ460949 KPJ10408.1 NWSH01000011 PCG80900.1 ODYU01003780 SOQ42965.1 KZ150052 PZC74388.1 JTDY01001496 KOB73733.1 BABH01004466 LJIG01016339 KRT80898.1 KQ971352 EFA06692.1 GEZM01068010 JAV67263.1 KB632281 ERL91329.1 APGK01054085 KB741247 ENN71992.1 BT128422 AEE63379.1 KK852527 KDR22010.1 GBYB01000296 JAG70063.1 KZ288322 PBC28225.1 NEVH01006994 PNF36065.1 CM000158 KRJ99442.1 AE013599 AY058750 AAG22280.2 AAL13979.1 BT003510 AAF58497.2 AAO39514.1
Proteomes
Interpro
ProteinModelPortal
C0KWE9
H9J3S1
B8Y9D1
A0A2H1VUX8
A0A2A4K5S1
A0A212F3W8
+ More
A0A3S2NUR2 A0A194PP62 A0A1E1WVG8 A0A2W1BIY9 A0A194QXZ1 A0A2A4KAY0 A0A2H1VQ46 A0A2W1BHR6 A0A0L7LDY1 H9J9C4 A0A0T6B0Q6 A0A1W4WK63 A0A1W4WW72 D6WTB7 A0A1Y1L0S3 U4UBP6 N6U0C7 J3JYU1 A0A067RN17 A0A0C9R0G7 A0A2A3E924 A0A2J7R5E8 A0A0R1DUE7 Q7K380 Q7JR08
A0A3S2NUR2 A0A194PP62 A0A1E1WVG8 A0A2W1BIY9 A0A194QXZ1 A0A2A4KAY0 A0A2H1VQ46 A0A2W1BHR6 A0A0L7LDY1 H9J9C4 A0A0T6B0Q6 A0A1W4WK63 A0A1W4WW72 D6WTB7 A0A1Y1L0S3 U4UBP6 N6U0C7 J3JYU1 A0A067RN17 A0A0C9R0G7 A0A2A3E924 A0A2J7R5E8 A0A0R1DUE7 Q7K380 Q7JR08
PDB
6FQP
E-value=1.76261e-26,
Score=296
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
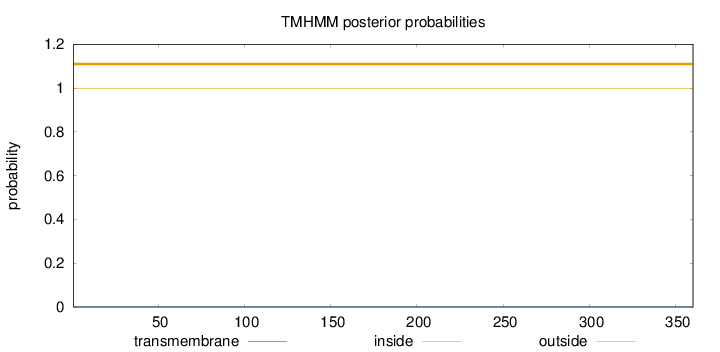
Length:
360
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00038
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00189
outside
1 - 360
Population Genetic Test Statistics
Pi
18.482025
Theta
19.665555
Tajima's D
0.051253
CLR
0.455862
CSRT
0.380930953452327
Interpretation
Uncertain
