Pre Gene Modal
BGIBMGA004159
Annotation
PREDICTED:_RNA_3'-terminal_phosphate_cyclase_[Papilio_machaon]
Full name
RNA 3'-terminal phosphate cyclase
Alternative Name
RNA terminal phosphate cyclase domain-containing protein 1
Location in the cell
Mitochondrial Reliability : 1.992
Sequence
CDS
ATGAATGAATTCTTGGAAATAGATGGGAGCATTTTGGAAGGGGGTGGTCAAGTACTCAGGAATTCAATATCATTAAGTGCAATTCTCGGAATACCTGTGCGTATTTTAAATATACGAGCTAACAGGAGCAAGCCAGGATTGGCACCCCAACATTTAAAAGGTGTAGAGCTAGTAGCAGAAATGTGTCAAGCCAAACTCAATGGTGCTCATATTCGGTCAACACAATTGGAGTTCAAACCTGGAAAGATTCGTGGAGGACATTATGTTGCTGATACACGCACAGCAGGTTCAATCAGTCTTTTACTCCAAGTGGCGCTACCCTGTGCAGTAATGGCGGACGGCCCCGTCACTCTGGAACTGAAGGGTGGCACCAATGCAGAGATGGCCCCGCAGATTGACTACATGACCGAGGTCTTTAAGCATGCAGTCAAACATTTTGGTGTGGATTTTAATCTAACCATACATAGAAGAGGGTATTTCCCGAAGGGTGGTGGTCACGTGACGGTGGAGGTGTCGCCCCTACGGCACGTGCAGGCCGCCGACGTGACGCGGGGGGGCGAAGTCGCCGCTGTGTACGGCTGGAGCTATGTAGCTGGCTGTCTGCCTATTAAAGTAAGATTGACGAGCAAGTTGTCCTCCGGCTACATTCTGGGAGGAGATGCTCTCGGCAAGAGAGGAGTGGATGCTGTGGAGGTCGGGAGGACGGCGGGAGAGATGCTCAGGGACGCTATCAGATCTGGTGCCTGCGTCGATAGTTATACTCAGGATCAATTGATTGTGTACATGGCGCTGGCGGACGGTCGGTCCAGGGTTCGAGTCGGTGAGATCACGCTGCACACGAGAACCGCAATACACTTCGCTGAACTCATCGCTAAGGCGAAGTTTAGCATACAGCCGGACGGGGATCAGAATGTGATCGAGTGCGACGGCGCAGGATACCACAATAGGAACCTGCCCGTCTAG
Protein
MNEFLEIDGSILEGGGQVLRNSISLSAILGIPVRILNIRANRSKPGLAPQHLKGVELVAEMCQAKLNGAHIRSTQLEFKPGKIRGGHYVADTRTAGSISLLLQVALPCAVMADGPVTLELKGGTNAEMAPQIDYMTEVFKHAVKHFGVDFNLTIHRRGYFPKGGGHVTVEVSPLRHVQAADVTRGGEVAAVYGWSYVAGCLPIKVRLTSKLSSGYILGGDALGKRGVDAVEVGRTAGEMLRDAIRSGACVDSYTQDQLIVYMALADGRSRVRVGEITLHTRTAIHFAELIAKAKFSIQPDGDQNVIECDGAGYHNRNLPV
Summary
Description
Catalyzes the conversion of 3'-phosphate to a 2',3'-cyclic phosphodiester at the end of RNA. The mechanism of action of the enzyme occurs in 3 steps: (A) adenylation of the enzyme by ATP; (B) transfer of adenylate to an RNA-N3'P to produce RNA-N3'PP5'A; (C) and attack of the adjacent 2'-hydroxyl on the 3'-phosphorus in the diester linkage to produce the cyclic end product. The biological role of this enzyme is unknown but it is likely to function in some aspects of cellular RNA processing (By similarity).
Catalytic Activity
(RNA)-3'-(3'-phospho-ribonucleoside) + ATP = (RNA)-3'-(2',3'-cyclophospho)-ribonucleoside + AMP + diphosphate
Subunit
Monomer.
Similarity
Belongs to the RNA 3'-terminal cyclase family. Type 1 subfamily.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Direct protein sequencing
Ligase
Nucleotide-binding
Nucleus
Reference proteome
Feature
chain RNA 3'-terminal phosphate cyclase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J3S2
A0A194R3J3
A0A194PMI4
A0A2H1VUZ5
A0A3S2NEQ1
A0A212F3W4
+ More
A0A2A4K5N6 H0Z446 A0A1Y1MPT0 K9J0A8 G3SNL8 A0A1W4XAG4 A0A3L8SQ40 H9ERH9 A0A2Y9SB92 A0A341BWI2 C3YTD9 A0A096MT18 A0A1U7UCB0 A0A383Z353 A0A2Y9E3L0 O00442 A0A340WJ97 A0A0D9S6M8 I2CT91 Q4R3J0 A0A2I3HIC5 H2N6M8 F7HIB0 G1T706 A0A2U3V1Z2 A0A2K5JVQ1 A0A250YGN2 K7CBD7 A0A1S3GDV7 F1S564 G3SGY2 A0A2K6EJ84 A0A2K6TX94 A0A1U7R0U0 A0A2Y9NYB7 A0A1A8AL30 A0A2P4T0D6 A0A1A8IBH0 A0A1L8DGH7 Q4T2M3 A0A3Q2W8V4 G5AYQ1 F7BTC6 Q2HJ88 A0A3B4F6H0 A0A3P8QM50 A0A3P9CPM6 Q5R7P3 A0A091D736 A8K8K1 A0A2I0UIE0 A0A3Q4G5M9 K7DHS2 A0A084VU98 A0A093GW58 A0A1A8REH3 A0A1B0CUP2 A0A3Q3H420 A0A2Y9IVS9 M3X9F8 A0A1A8LPC6 A0A226PPU1 F7GBC6 A0A1A8E1J5 Q9D7H3 A0A2M3ZC87 R7TZB6 A0A087R9M0 A0A2M3ZC55 A0A2K6NPQ1 L5K5R9 U6CW30 G9KM20 Q91YL8 A0A384CN37 A0A3Q7VMB8 Q5BLJ4 A0A2K5D2I6 A0A2I2ZH37 H2TBR3 F7H6I6 A0A067RIS5 Q68FS8 A0A2K5PXC9 W5JFK2 I3JLM5 A0A1A8FQN7 H3CGK0 A0A3Q7P322 A0A182FE17 A0A3Q7SBD4 V8P0D2 V9KVW2 A0A2U3W0P3
A0A2A4K5N6 H0Z446 A0A1Y1MPT0 K9J0A8 G3SNL8 A0A1W4XAG4 A0A3L8SQ40 H9ERH9 A0A2Y9SB92 A0A341BWI2 C3YTD9 A0A096MT18 A0A1U7UCB0 A0A383Z353 A0A2Y9E3L0 O00442 A0A340WJ97 A0A0D9S6M8 I2CT91 Q4R3J0 A0A2I3HIC5 H2N6M8 F7HIB0 G1T706 A0A2U3V1Z2 A0A2K5JVQ1 A0A250YGN2 K7CBD7 A0A1S3GDV7 F1S564 G3SGY2 A0A2K6EJ84 A0A2K6TX94 A0A1U7R0U0 A0A2Y9NYB7 A0A1A8AL30 A0A2P4T0D6 A0A1A8IBH0 A0A1L8DGH7 Q4T2M3 A0A3Q2W8V4 G5AYQ1 F7BTC6 Q2HJ88 A0A3B4F6H0 A0A3P8QM50 A0A3P9CPM6 Q5R7P3 A0A091D736 A8K8K1 A0A2I0UIE0 A0A3Q4G5M9 K7DHS2 A0A084VU98 A0A093GW58 A0A1A8REH3 A0A1B0CUP2 A0A3Q3H420 A0A2Y9IVS9 M3X9F8 A0A1A8LPC6 A0A226PPU1 F7GBC6 A0A1A8E1J5 Q9D7H3 A0A2M3ZC87 R7TZB6 A0A087R9M0 A0A2M3ZC55 A0A2K6NPQ1 L5K5R9 U6CW30 G9KM20 Q91YL8 A0A384CN37 A0A3Q7VMB8 Q5BLJ4 A0A2K5D2I6 A0A2I2ZH37 H2TBR3 F7H6I6 A0A067RIS5 Q68FS8 A0A2K5PXC9 W5JFK2 I3JLM5 A0A1A8FQN7 H3CGK0 A0A3Q7P322 A0A182FE17 A0A3Q7SBD4 V8P0D2 V9KVW2 A0A2U3W0P3
EC Number
6.5.1.4
Pubmed
19121390
26354079
22118469
20360741
28004739
30282656
+ More
25319552 18563158 9184239 16710414 15489334 2199762 21269460 17431167 21993624 28087693 30723633 15496914 21993625 19892987 25186727 24438588 17975172 24621616 17495919 16141072 21183079 23254933 25362486 23258410 23236062 24813606 21551351 25243066 24845553 15057822 15632090 20920257 23761445 24297900 24402279
25319552 18563158 9184239 16710414 15489334 2199762 21269460 17431167 21993624 28087693 30723633 15496914 21993625 19892987 25186727 24438588 17975172 24621616 17495919 16141072 21183079 23254933 25362486 23258410 23236062 24813606 21551351 25243066 24845553 15057822 15632090 20920257 23761445 24297900 24402279
EMBL
BABH01028864
KQ460949
KPJ10411.1
KQ459598
KPI94532.1
ODYU01004598
+ More
SOQ44649.1 RSAL01000070 RVE49192.1 AGBW02010485 OWR48421.1 NWSH01000119 PCG79379.1 ABQF01035310 GEZM01028102 JAV86465.1 GABZ01006661 JAA46864.1 QUSF01000010 RLW05882.1 JU321232 JU473295 JV048550 AFE64988.1 AFH30099.1 AFI38621.1 GG666551 EEN56506.1 AHZZ02016465 Y11651 Y11652 AL445928 AL663111 CH471097 BC012604 AQIB01085936 JV489142 AFJ70176.1 AB179276 ADFV01190039 ABGA01284966 ABGA01284967 ABGA01284968 NDHI03003423 PNJ55251.1 JSUE03002012 JSUE03002013 JSUE03002014 AAGW02001261 GFFW01002027 JAV42761.1 GABC01004205 GABF01005938 NBAG03000257 JAA07133.1 JAA16207.1 PNI59024.1 AEMK02000025 DQIR01126939 DQIR01205313 DQIR01215804 DQIR01293392 DQIR01323035 HDA82415.1 HDB60790.1 CABD030004840 HADY01016908 SBP55393.1 PPHD01014024 POI29800.1 HAED01007977 HAEE01012916 SBQ94189.1 GFDF01008521 JAV05563.1 CAAE01010234 CAF92859.1 JH167580 GEBF01005144 EHB02162.1 JAN98488.1 BC113251 CR860069 KN123067 KFO26887.1 AK292366 BAF85055.1 KZ505741 PKU45813.1 GABD01008128 GABE01010837 JAA24972.1 JAA33902.1 ATLV01016710 KE525103 KFB41542.1 KL216678 KFV71029.1 HAEH01015093 HAEI01008218 SBS04436.1 AJWK01029481 AANG04003279 HAEF01008217 SBR46176.1 AWGT02000036 OXB81500.1 HADZ01010828 HAEA01011567 SBQ40047.1 AK009245 AK152129 AK159961 CH466532 GGFM01005403 MBW26154.1 AMQN01001957 KB307198 ELT99109.1 KL226226 KFM10174.1 GGFM01005338 MBW26089.1 KB031037 ELK05828.1 HAAF01002845 CCP74671.1 AEYP01085891 JP017351 AES05949.1 BC016519 AAH16519.1 BC090403 AAH90403.1 GAMT01001913 GAMT01001912 GAMR01009539 GAMR01009538 GAMQ01003049 GAMQ01003048 GAMQ01003047 GAMP01000838 GAMP01000837 JAB09948.1 JAB24393.1 JAB38802.1 JAB51917.1 KK852607 KDR20351.1 AC119448 BC079374 CH473952 AAH79374.1 EDL82031.1 ADMH02001288 ETN63152.1 AERX01020296 HAEB01014636 HAEC01005441 SBQ61163.1 AZIM01001230 ETE67641.1 JW870279 AFP02797.1
SOQ44649.1 RSAL01000070 RVE49192.1 AGBW02010485 OWR48421.1 NWSH01000119 PCG79379.1 ABQF01035310 GEZM01028102 JAV86465.1 GABZ01006661 JAA46864.1 QUSF01000010 RLW05882.1 JU321232 JU473295 JV048550 AFE64988.1 AFH30099.1 AFI38621.1 GG666551 EEN56506.1 AHZZ02016465 Y11651 Y11652 AL445928 AL663111 CH471097 BC012604 AQIB01085936 JV489142 AFJ70176.1 AB179276 ADFV01190039 ABGA01284966 ABGA01284967 ABGA01284968 NDHI03003423 PNJ55251.1 JSUE03002012 JSUE03002013 JSUE03002014 AAGW02001261 GFFW01002027 JAV42761.1 GABC01004205 GABF01005938 NBAG03000257 JAA07133.1 JAA16207.1 PNI59024.1 AEMK02000025 DQIR01126939 DQIR01205313 DQIR01215804 DQIR01293392 DQIR01323035 HDA82415.1 HDB60790.1 CABD030004840 HADY01016908 SBP55393.1 PPHD01014024 POI29800.1 HAED01007977 HAEE01012916 SBQ94189.1 GFDF01008521 JAV05563.1 CAAE01010234 CAF92859.1 JH167580 GEBF01005144 EHB02162.1 JAN98488.1 BC113251 CR860069 KN123067 KFO26887.1 AK292366 BAF85055.1 KZ505741 PKU45813.1 GABD01008128 GABE01010837 JAA24972.1 JAA33902.1 ATLV01016710 KE525103 KFB41542.1 KL216678 KFV71029.1 HAEH01015093 HAEI01008218 SBS04436.1 AJWK01029481 AANG04003279 HAEF01008217 SBR46176.1 AWGT02000036 OXB81500.1 HADZ01010828 HAEA01011567 SBQ40047.1 AK009245 AK152129 AK159961 CH466532 GGFM01005403 MBW26154.1 AMQN01001957 KB307198 ELT99109.1 KL226226 KFM10174.1 GGFM01005338 MBW26089.1 KB031037 ELK05828.1 HAAF01002845 CCP74671.1 AEYP01085891 JP017351 AES05949.1 BC016519 AAH16519.1 BC090403 AAH90403.1 GAMT01001913 GAMT01001912 GAMR01009539 GAMR01009538 GAMQ01003049 GAMQ01003048 GAMQ01003047 GAMP01000838 GAMP01000837 JAB09948.1 JAB24393.1 JAB38802.1 JAB51917.1 KK852607 KDR20351.1 AC119448 BC079374 CH473952 AAH79374.1 EDL82031.1 ADMH02001288 ETN63152.1 AERX01020296 HAEB01014636 HAEC01005441 SBQ61163.1 AZIM01001230 ETE67641.1 JW870279 AFP02797.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000218220
+ More
UP000007754 UP000007646 UP000192223 UP000276834 UP000248484 UP000252040 UP000001554 UP000028761 UP000189704 UP000261681 UP000248480 UP000005640 UP000265300 UP000029965 UP000233100 UP000001073 UP000001595 UP000006718 UP000001811 UP000245320 UP000233080 UP000081671 UP000008227 UP000001519 UP000233160 UP000233220 UP000189706 UP000248483 UP000264840 UP000006813 UP000002281 UP000009136 UP000261460 UP000265100 UP000265160 UP000028990 UP000261580 UP000030765 UP000053875 UP000092461 UP000261660 UP000248482 UP000011712 UP000198419 UP000002280 UP000000589 UP000014760 UP000053286 UP000233200 UP000010552 UP000000715 UP000261680 UP000291021 UP000286642 UP000233020 UP000005226 UP000008225 UP000027135 UP000002494 UP000233040 UP000000673 UP000005207 UP000007303 UP000286641 UP000069272 UP000286640 UP000245340
UP000007754 UP000007646 UP000192223 UP000276834 UP000248484 UP000252040 UP000001554 UP000028761 UP000189704 UP000261681 UP000248480 UP000005640 UP000265300 UP000029965 UP000233100 UP000001073 UP000001595 UP000006718 UP000001811 UP000245320 UP000233080 UP000081671 UP000008227 UP000001519 UP000233160 UP000233220 UP000189706 UP000248483 UP000264840 UP000006813 UP000002281 UP000009136 UP000261460 UP000265100 UP000265160 UP000028990 UP000261580 UP000030765 UP000053875 UP000092461 UP000261660 UP000248482 UP000011712 UP000198419 UP000002280 UP000000589 UP000014760 UP000053286 UP000233200 UP000010552 UP000000715 UP000261680 UP000291021 UP000286642 UP000233020 UP000005226 UP000008225 UP000027135 UP000002494 UP000233040 UP000000673 UP000005207 UP000007303 UP000286641 UP000069272 UP000286640 UP000245340
Interpro
Gene 3D
ProteinModelPortal
H9J3S2
A0A194R3J3
A0A194PMI4
A0A2H1VUZ5
A0A3S2NEQ1
A0A212F3W4
+ More
A0A2A4K5N6 H0Z446 A0A1Y1MPT0 K9J0A8 G3SNL8 A0A1W4XAG4 A0A3L8SQ40 H9ERH9 A0A2Y9SB92 A0A341BWI2 C3YTD9 A0A096MT18 A0A1U7UCB0 A0A383Z353 A0A2Y9E3L0 O00442 A0A340WJ97 A0A0D9S6M8 I2CT91 Q4R3J0 A0A2I3HIC5 H2N6M8 F7HIB0 G1T706 A0A2U3V1Z2 A0A2K5JVQ1 A0A250YGN2 K7CBD7 A0A1S3GDV7 F1S564 G3SGY2 A0A2K6EJ84 A0A2K6TX94 A0A1U7R0U0 A0A2Y9NYB7 A0A1A8AL30 A0A2P4T0D6 A0A1A8IBH0 A0A1L8DGH7 Q4T2M3 A0A3Q2W8V4 G5AYQ1 F7BTC6 Q2HJ88 A0A3B4F6H0 A0A3P8QM50 A0A3P9CPM6 Q5R7P3 A0A091D736 A8K8K1 A0A2I0UIE0 A0A3Q4G5M9 K7DHS2 A0A084VU98 A0A093GW58 A0A1A8REH3 A0A1B0CUP2 A0A3Q3H420 A0A2Y9IVS9 M3X9F8 A0A1A8LPC6 A0A226PPU1 F7GBC6 A0A1A8E1J5 Q9D7H3 A0A2M3ZC87 R7TZB6 A0A087R9M0 A0A2M3ZC55 A0A2K6NPQ1 L5K5R9 U6CW30 G9KM20 Q91YL8 A0A384CN37 A0A3Q7VMB8 Q5BLJ4 A0A2K5D2I6 A0A2I2ZH37 H2TBR3 F7H6I6 A0A067RIS5 Q68FS8 A0A2K5PXC9 W5JFK2 I3JLM5 A0A1A8FQN7 H3CGK0 A0A3Q7P322 A0A182FE17 A0A3Q7SBD4 V8P0D2 V9KVW2 A0A2U3W0P3
A0A2A4K5N6 H0Z446 A0A1Y1MPT0 K9J0A8 G3SNL8 A0A1W4XAG4 A0A3L8SQ40 H9ERH9 A0A2Y9SB92 A0A341BWI2 C3YTD9 A0A096MT18 A0A1U7UCB0 A0A383Z353 A0A2Y9E3L0 O00442 A0A340WJ97 A0A0D9S6M8 I2CT91 Q4R3J0 A0A2I3HIC5 H2N6M8 F7HIB0 G1T706 A0A2U3V1Z2 A0A2K5JVQ1 A0A250YGN2 K7CBD7 A0A1S3GDV7 F1S564 G3SGY2 A0A2K6EJ84 A0A2K6TX94 A0A1U7R0U0 A0A2Y9NYB7 A0A1A8AL30 A0A2P4T0D6 A0A1A8IBH0 A0A1L8DGH7 Q4T2M3 A0A3Q2W8V4 G5AYQ1 F7BTC6 Q2HJ88 A0A3B4F6H0 A0A3P8QM50 A0A3P9CPM6 Q5R7P3 A0A091D736 A8K8K1 A0A2I0UIE0 A0A3Q4G5M9 K7DHS2 A0A084VU98 A0A093GW58 A0A1A8REH3 A0A1B0CUP2 A0A3Q3H420 A0A2Y9IVS9 M3X9F8 A0A1A8LPC6 A0A226PPU1 F7GBC6 A0A1A8E1J5 Q9D7H3 A0A2M3ZC87 R7TZB6 A0A087R9M0 A0A2M3ZC55 A0A2K6NPQ1 L5K5R9 U6CW30 G9KM20 Q91YL8 A0A384CN37 A0A3Q7VMB8 Q5BLJ4 A0A2K5D2I6 A0A2I2ZH37 H2TBR3 F7H6I6 A0A067RIS5 Q68FS8 A0A2K5PXC9 W5JFK2 I3JLM5 A0A1A8FQN7 H3CGK0 A0A3Q7P322 A0A182FE17 A0A3Q7SBD4 V8P0D2 V9KVW2 A0A2U3W0P3
PDB
4O8J
E-value=7.15259e-47,
Score=471
Ontologies
GO
PANTHER
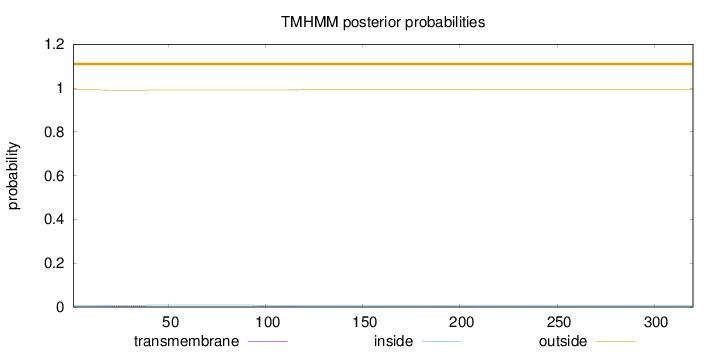
Topology
Subcellular location
Length:
320
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.127
Exp number, first 60 AAs:
0.0853800000000001
Total prob of N-in:
0.00776
outside
1 - 320
Population Genetic Test Statistics
Pi
19.965514
Theta
21.423504
Tajima's D
-0.684316
CLR
1.127381
CSRT
0.200589970501475
Interpretation
Uncertain
