Gene
KWMTBOMO11734 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004191
Annotation
PREDICTED:_zinc_finger_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.643
Sequence
CDS
ATGAGTTCAAGCGTTTGCTACAAGTGCAACCGGACCGGGCATTTCGCGCGCGAATGCACGCAGGGGGGCGTGGTGTCGCGGGATTCCGGTTTCAATCGGCAACGTGAGAAGTGCTTCAAGTGCAACCGCACAGGACACTTTGCGAGGGATTGCAAGGAAGAGGCTGACCGTTGCTACAGATGTAACGGCACTGGGCACATAGCGCGCGAGTGCGCACAGAGCCCGGACGAGCCGTCATGTTACAACTGCAACAAGACGGGCCACATCGCACGGAACTGTCCCGAGGGGGGGCGGGAGTCTGCGACGCAGACCTGCTATAACTGCAACAAGTCCGGCCACATCTCCCGCAACTGTCCCGACGGCACCAAGACGTGCTACGTGTGCGGCAAGCCCGGCCACATCTCGCGCGAGTGCGACGAGGCGCGGAACTAG
Protein
MSSSVCYKCNRTGHFARECTQGGVVSRDSGFNRQREKCFKCNRTGHFARDCKEEADRCYRCNGTGHIARECAQSPDEPSCYNCNKTGHIARNCPEGGRESATQTCYNCNKSGHISRNCPDGTKTCYVCGKPGHISRECDEARN
Summary
Uniprot
Q3L234
A0A3S2TL92
A0A2A4K6G8
A0A2A4K542
A0A0K8SLJ4
A0A2H1V338
+ More
I4DIY6 A0A194R3K3 A0A194PTS6 I4DM61 A0A212F3W1 A0A1E1WT25 A0A0L7LQN1 A0A1B0CTD1 A0A1L8DSS2 B7U4P9 A0A1B6ECJ2 A0A1B6F381 A0A1W4WYI3 A0A1B6KW45 D1FPM1 A0A139WF77 A0A067RA31 R4UK13 A0A151IWZ0 A0A195EW39 E2BYY6 A0A158NNV4 E2A8B4 A0A151ICA4 R4WNK7 A0A182J707 A0A084VN92 A0A0T6ATF8 A0A2J7QCN2 A0A2J7QMS3 T1E829 A0A2M4CSS5 A0A2M3YXX9 A0A2M4A094 A0A2M4C237 Q7PNE6 A0A3L8DNL0 A0A0M8ZWY1 A0A067QNY2 A0A1D1XQS6 W5JKG9 T1E802 J9HTV7 T1PFN3 A0A2A3E706 A0A0L7RFS0 V9IKH5 A0A087ZVA0 K7IM87 A0A224XVP6 A0A0V0G2P5 A0A069DNX6 R4FNM1 A0A023F9L7 A0A1Q3F3Q2 T1DIY4 A0A1Q3F5A4 A0A1Q3F6W3 T1H7E5 A0A182UXT3 A0A1I8PVL0 T1IWB3 N6UC67 A0A182L1M3 A0A2R7WMY2 D3TRZ3 B0W4N4 A0A182F6C6 A0A0P4VYS6 A0A1L8EEM1 A0A1Q3EZP0 W8CDG6 A0A0P4WXY5 A0A0A9YBN6 A0A0P4WHY6 A0A182Y043 A0A0K8WLI5 A0A034WW73 A0A0C9RXL0 A0A0A1WN46 A0A0P4WQB0 C6ZQR4 A0A0P4VTP0 A0A0N8P0F2 A0A1S3CYV4 F4WVA6 A0A195BRX5 A0A0J7NPD9 A0A0K8TT64 A0A3B0J2J9 I7M5V3 A0A1S3D2Q5 A0A3R7SSU7
I4DIY6 A0A194R3K3 A0A194PTS6 I4DM61 A0A212F3W1 A0A1E1WT25 A0A0L7LQN1 A0A1B0CTD1 A0A1L8DSS2 B7U4P9 A0A1B6ECJ2 A0A1B6F381 A0A1W4WYI3 A0A1B6KW45 D1FPM1 A0A139WF77 A0A067RA31 R4UK13 A0A151IWZ0 A0A195EW39 E2BYY6 A0A158NNV4 E2A8B4 A0A151ICA4 R4WNK7 A0A182J707 A0A084VN92 A0A0T6ATF8 A0A2J7QCN2 A0A2J7QMS3 T1E829 A0A2M4CSS5 A0A2M3YXX9 A0A2M4A094 A0A2M4C237 Q7PNE6 A0A3L8DNL0 A0A0M8ZWY1 A0A067QNY2 A0A1D1XQS6 W5JKG9 T1E802 J9HTV7 T1PFN3 A0A2A3E706 A0A0L7RFS0 V9IKH5 A0A087ZVA0 K7IM87 A0A224XVP6 A0A0V0G2P5 A0A069DNX6 R4FNM1 A0A023F9L7 A0A1Q3F3Q2 T1DIY4 A0A1Q3F5A4 A0A1Q3F6W3 T1H7E5 A0A182UXT3 A0A1I8PVL0 T1IWB3 N6UC67 A0A182L1M3 A0A2R7WMY2 D3TRZ3 B0W4N4 A0A182F6C6 A0A0P4VYS6 A0A1L8EEM1 A0A1Q3EZP0 W8CDG6 A0A0P4WXY5 A0A0A9YBN6 A0A0P4WHY6 A0A182Y043 A0A0K8WLI5 A0A034WW73 A0A0C9RXL0 A0A0A1WN46 A0A0P4WQB0 C6ZQR4 A0A0P4VTP0 A0A0N8P0F2 A0A1S3CYV4 F4WVA6 A0A195BRX5 A0A0J7NPD9 A0A0K8TT64 A0A3B0J2J9 I7M5V3 A0A1S3D2Q5 A0A3R7SSU7
Pubmed
19121390
22651552
26354079
22118469
26227816
20441151
+ More
18362917 19820115 24845553 20798317 21347285 23691247 24438588 12364791 14747013 17210077 30249741 20920257 23761445 17510324 25315136 20075255 26334808 25474469 24330624 23537049 20966253 20353571 27129103 24495485 25401762 25244985 25348373 25830018 20496585 17994087 21719571 26369729 15632085
18362917 19820115 24845553 20798317 21347285 23691247 24438588 12364791 14747013 17210077 30249741 20920257 23761445 17510324 25315136 20075255 26334808 25474469 24330624 23537049 20966253 20353571 27129103 24495485 25401762 25244985 25348373 25830018 20496585 17994087 21719571 26369729 15632085
EMBL
BABH01028843
AY753659
AAX09282.1
RSAL01000070
RVE49186.1
NWSH01000119
+ More
PCG79363.1 PCG79361.1 GBRD01011617 JAG54207.1 ODYU01000451 SOQ35258.1 AK401254 BAM17876.1 KQ460949 KPJ10421.1 KQ459598 KPI94540.1 AK402379 BAM19001.1 AGBW02010485 OWR48414.1 GDQN01010323 GDQN01000861 GDQN01000552 JAT80731.1 JAT90193.1 JAT90502.1 JTDY01000300 KOB77808.1 AJWK01027434 GFDF01004562 JAV09522.1 FJ430683 ACJ50597.1 GEDC01001657 JAS35641.1 GECZ01026737 GECZ01025050 GECZ01001361 JAS43032.1 JAS44719.1 JAS68408.1 GEBQ01024285 JAT15692.1 EZ419771 ACY69944.1 KQ971352 KYB26648.1 KK852643 KDR19567.1 KC571915 AGM32414.1 KQ980843 KYN12293.1 KQ981953 KYN32453.1 GL451531 EFN79104.1 ADTU01021805 GL437526 EFN70327.1 KQ978062 KYM97714.1 AK417226 BAN20441.1 ATLV01014730 KE524984 KFB39436.1 LJIG01022865 KRT78326.1 NEVH01016289 PNF26333.1 NEVH01013204 PNF29882.1 GAMD01003108 JAA98482.1 GGFL01004053 MBW68231.1 GGFM01000363 MBW21114.1 GGFK01000913 MBW34234.1 GGFJ01010236 MBW59377.1 AAAB01008964 EAA12317.4 QOIP01000006 RLU21876.1 KQ435830 KOX71736.1 KK853183 KDR10130.1 GDJX01023206 JAT44730.1 ADMH02000936 ETN64631.1 GAMD01003178 JAA98412.1 CH478212 EJY58088.1 KA647469 AFP62098.1 KZ288348 PBC27495.1 KQ414598 KOC69832.1 JR048970 AEY60819.1 GFTR01002508 JAW13918.1 GECL01003721 JAP02403.1 GBGD01003349 JAC85540.1 ACPB03019911 GAHY01001234 JAA76276.1 GBBI01000516 GEMB01000692 JAC18196.1 JAS02443.1 GFDL01012853 JAV22192.1 GALA01000666 JAA94186.1 GFDL01012336 JAV22709.1 GFDL01011745 JAV23300.1 JH431612 APGK01029404 APGK01029405 KB740734 ENN79255.1 KK855120 PTY20976.1 EZ424195 ADD20471.1 DS231838 EDS33879.1 GDKW01001889 JAI54706.1 GFDG01001658 JAV17141.1 GFDL01014286 JAV20759.1 GAMC01001981 JAC04575.1 GDRN01050044 JAI66491.1 GBHO01025045 GBHO01014040 JAG18559.1 JAG29564.1 GDRN01050045 JAI66490.1 GDHF01017742 GDHF01000308 JAI34572.1 JAI52006.1 GAKP01000899 JAC58053.1 GBYB01013910 JAG83677.1 GBXI01014025 JAD00267.1 GDRN01050046 GDRN01050043 JAI66492.1 EZ114865 ACU30918.1 GDKW01001250 JAI55345.1 CH902619 KPU76956.1 GL888384 EGI61872.1 KQ976417 KYM89782.1 LBMM01002789 KMQ94360.1 GDAI01000508 JAI17095.1 OUUW01000001 SPP73293.1 CM000071 EAL26521.1 QCYY01002023 ROT73514.1
PCG79363.1 PCG79361.1 GBRD01011617 JAG54207.1 ODYU01000451 SOQ35258.1 AK401254 BAM17876.1 KQ460949 KPJ10421.1 KQ459598 KPI94540.1 AK402379 BAM19001.1 AGBW02010485 OWR48414.1 GDQN01010323 GDQN01000861 GDQN01000552 JAT80731.1 JAT90193.1 JAT90502.1 JTDY01000300 KOB77808.1 AJWK01027434 GFDF01004562 JAV09522.1 FJ430683 ACJ50597.1 GEDC01001657 JAS35641.1 GECZ01026737 GECZ01025050 GECZ01001361 JAS43032.1 JAS44719.1 JAS68408.1 GEBQ01024285 JAT15692.1 EZ419771 ACY69944.1 KQ971352 KYB26648.1 KK852643 KDR19567.1 KC571915 AGM32414.1 KQ980843 KYN12293.1 KQ981953 KYN32453.1 GL451531 EFN79104.1 ADTU01021805 GL437526 EFN70327.1 KQ978062 KYM97714.1 AK417226 BAN20441.1 ATLV01014730 KE524984 KFB39436.1 LJIG01022865 KRT78326.1 NEVH01016289 PNF26333.1 NEVH01013204 PNF29882.1 GAMD01003108 JAA98482.1 GGFL01004053 MBW68231.1 GGFM01000363 MBW21114.1 GGFK01000913 MBW34234.1 GGFJ01010236 MBW59377.1 AAAB01008964 EAA12317.4 QOIP01000006 RLU21876.1 KQ435830 KOX71736.1 KK853183 KDR10130.1 GDJX01023206 JAT44730.1 ADMH02000936 ETN64631.1 GAMD01003178 JAA98412.1 CH478212 EJY58088.1 KA647469 AFP62098.1 KZ288348 PBC27495.1 KQ414598 KOC69832.1 JR048970 AEY60819.1 GFTR01002508 JAW13918.1 GECL01003721 JAP02403.1 GBGD01003349 JAC85540.1 ACPB03019911 GAHY01001234 JAA76276.1 GBBI01000516 GEMB01000692 JAC18196.1 JAS02443.1 GFDL01012853 JAV22192.1 GALA01000666 JAA94186.1 GFDL01012336 JAV22709.1 GFDL01011745 JAV23300.1 JH431612 APGK01029404 APGK01029405 KB740734 ENN79255.1 KK855120 PTY20976.1 EZ424195 ADD20471.1 DS231838 EDS33879.1 GDKW01001889 JAI54706.1 GFDG01001658 JAV17141.1 GFDL01014286 JAV20759.1 GAMC01001981 JAC04575.1 GDRN01050044 JAI66491.1 GBHO01025045 GBHO01014040 JAG18559.1 JAG29564.1 GDRN01050045 JAI66490.1 GDHF01017742 GDHF01000308 JAI34572.1 JAI52006.1 GAKP01000899 JAC58053.1 GBYB01013910 JAG83677.1 GBXI01014025 JAD00267.1 GDRN01050046 GDRN01050043 JAI66492.1 EZ114865 ACU30918.1 GDKW01001250 JAI55345.1 CH902619 KPU76956.1 GL888384 EGI61872.1 KQ976417 KYM89782.1 LBMM01002789 KMQ94360.1 GDAI01000508 JAI17095.1 OUUW01000001 SPP73293.1 CM000071 EAL26521.1 QCYY01002023 ROT73514.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000092461 UP000192223 UP000007266 UP000027135 UP000078492 UP000078541 UP000008237 UP000005205 UP000000311 UP000078542 UP000075880 UP000030765 UP000235965 UP000007062 UP000279307 UP000053105 UP000000673 UP000008820 UP000095301 UP000242457 UP000053825 UP000005203 UP000002358 UP000015103 UP000015102 UP000075903 UP000095300 UP000019118 UP000075882 UP000002320 UP000069272 UP000076408 UP000007801 UP000079169 UP000007755 UP000078540 UP000036403 UP000268350 UP000001819 UP000283509
UP000037510 UP000092461 UP000192223 UP000007266 UP000027135 UP000078492 UP000078541 UP000008237 UP000005205 UP000000311 UP000078542 UP000075880 UP000030765 UP000235965 UP000007062 UP000279307 UP000053105 UP000000673 UP000008820 UP000095301 UP000242457 UP000053825 UP000005203 UP000002358 UP000015103 UP000015102 UP000075903 UP000095300 UP000019118 UP000075882 UP000002320 UP000069272 UP000076408 UP000007801 UP000079169 UP000007755 UP000078540 UP000036403 UP000268350 UP000001819 UP000283509
PRIDE
Interpro
SUPFAM
SSF57756
SSF57756
ProteinModelPortal
Q3L234
A0A3S2TL92
A0A2A4K6G8
A0A2A4K542
A0A0K8SLJ4
A0A2H1V338
+ More
I4DIY6 A0A194R3K3 A0A194PTS6 I4DM61 A0A212F3W1 A0A1E1WT25 A0A0L7LQN1 A0A1B0CTD1 A0A1L8DSS2 B7U4P9 A0A1B6ECJ2 A0A1B6F381 A0A1W4WYI3 A0A1B6KW45 D1FPM1 A0A139WF77 A0A067RA31 R4UK13 A0A151IWZ0 A0A195EW39 E2BYY6 A0A158NNV4 E2A8B4 A0A151ICA4 R4WNK7 A0A182J707 A0A084VN92 A0A0T6ATF8 A0A2J7QCN2 A0A2J7QMS3 T1E829 A0A2M4CSS5 A0A2M3YXX9 A0A2M4A094 A0A2M4C237 Q7PNE6 A0A3L8DNL0 A0A0M8ZWY1 A0A067QNY2 A0A1D1XQS6 W5JKG9 T1E802 J9HTV7 T1PFN3 A0A2A3E706 A0A0L7RFS0 V9IKH5 A0A087ZVA0 K7IM87 A0A224XVP6 A0A0V0G2P5 A0A069DNX6 R4FNM1 A0A023F9L7 A0A1Q3F3Q2 T1DIY4 A0A1Q3F5A4 A0A1Q3F6W3 T1H7E5 A0A182UXT3 A0A1I8PVL0 T1IWB3 N6UC67 A0A182L1M3 A0A2R7WMY2 D3TRZ3 B0W4N4 A0A182F6C6 A0A0P4VYS6 A0A1L8EEM1 A0A1Q3EZP0 W8CDG6 A0A0P4WXY5 A0A0A9YBN6 A0A0P4WHY6 A0A182Y043 A0A0K8WLI5 A0A034WW73 A0A0C9RXL0 A0A0A1WN46 A0A0P4WQB0 C6ZQR4 A0A0P4VTP0 A0A0N8P0F2 A0A1S3CYV4 F4WVA6 A0A195BRX5 A0A0J7NPD9 A0A0K8TT64 A0A3B0J2J9 I7M5V3 A0A1S3D2Q5 A0A3R7SSU7
I4DIY6 A0A194R3K3 A0A194PTS6 I4DM61 A0A212F3W1 A0A1E1WT25 A0A0L7LQN1 A0A1B0CTD1 A0A1L8DSS2 B7U4P9 A0A1B6ECJ2 A0A1B6F381 A0A1W4WYI3 A0A1B6KW45 D1FPM1 A0A139WF77 A0A067RA31 R4UK13 A0A151IWZ0 A0A195EW39 E2BYY6 A0A158NNV4 E2A8B4 A0A151ICA4 R4WNK7 A0A182J707 A0A084VN92 A0A0T6ATF8 A0A2J7QCN2 A0A2J7QMS3 T1E829 A0A2M4CSS5 A0A2M3YXX9 A0A2M4A094 A0A2M4C237 Q7PNE6 A0A3L8DNL0 A0A0M8ZWY1 A0A067QNY2 A0A1D1XQS6 W5JKG9 T1E802 J9HTV7 T1PFN3 A0A2A3E706 A0A0L7RFS0 V9IKH5 A0A087ZVA0 K7IM87 A0A224XVP6 A0A0V0G2P5 A0A069DNX6 R4FNM1 A0A023F9L7 A0A1Q3F3Q2 T1DIY4 A0A1Q3F5A4 A0A1Q3F6W3 T1H7E5 A0A182UXT3 A0A1I8PVL0 T1IWB3 N6UC67 A0A182L1M3 A0A2R7WMY2 D3TRZ3 B0W4N4 A0A182F6C6 A0A0P4VYS6 A0A1L8EEM1 A0A1Q3EZP0 W8CDG6 A0A0P4WXY5 A0A0A9YBN6 A0A0P4WHY6 A0A182Y043 A0A0K8WLI5 A0A034WW73 A0A0C9RXL0 A0A0A1WN46 A0A0P4WQB0 C6ZQR4 A0A0P4VTP0 A0A0N8P0F2 A0A1S3CYV4 F4WVA6 A0A195BRX5 A0A0J7NPD9 A0A0K8TT64 A0A3B0J2J9 I7M5V3 A0A1S3D2Q5 A0A3R7SSU7
PDB
5O2U
E-value=9.09209e-06,
Score=111
Ontologies
GO
PANTHER
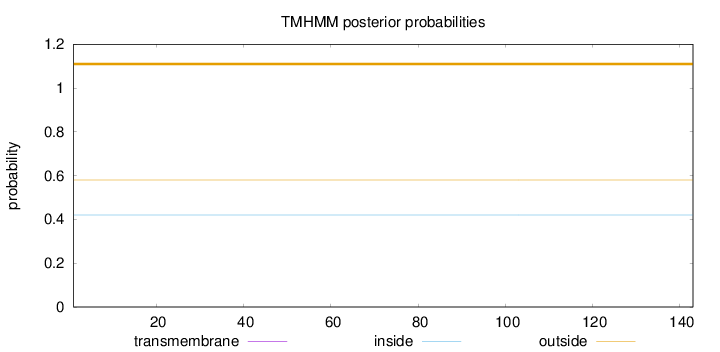
Topology
Length:
143
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.42025
outside
1 - 143
Population Genetic Test Statistics
Pi
27.694719
Theta
24.408841
Tajima's D
0.447577
CLR
1.986752
CSRT
0.498025098745063
Interpretation
Uncertain
