Gene
KWMTBOMO11731 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004165
Annotation
translation_elongation_factor_2_isoform_2_[Bombyx_mori]
Full name
Translation elongation factor 2
+ More
Elongation factor 2
Elongation factor 2
Location in the cell
Cytoplasmic Reliability : 4.299
Sequence
CDS
ATGGTGAATTTCACGGTAGACGAGATCCGTGGGATGATGGACAAGAAGCGGAATATCCGCAACATGTCTGTGATCGCCCACGTCGATCACGGCAAGTCAACCCTCACGGACTCGTTGGTTTCCAAGGCCGGTATCATTGCTGGTGCGAGAGCCGGAGAGACCCGTTTCACTGACACGCGTAAGGACGAACAAGACCGTTGCATCACCATTAAATCTACGGCCATCTCTATGTTCTTCGAGCTTGAAGAGAAAGATTTAGTATTCATCACAAACCCTGACCAGCGTGAAAAGAGTGAGAAAGGTTTCTTGATCAACTTGATTGACTCACCTGGACACGTTGATTTCTCTTCTGAAGTAACAGCTGCACTCCGTGTCACTGATGGAGCCCTTGTGGTTGTTGACTGTGTGTCTGGTGTGTGTGTACAAACTGAAACAGTACTGCGTCAGGCTATTGCCGAGCGCATCAAGCCTATTCTGTTCATGAACAAAATGGACCGTGCTCTTCTTGAGCTCCAACTTGAAGCTGAAGAACTATACCAGACGTTCCAGCGTATTGTAGAAAATGTTAACGTCATTATAGCCACATATAACGATGATGGTGGTCCCATGGGTGAGGTGCGTGTCGACCCTAGCAAGGGCTCTGTTGGTTTCGGGTCTGGTCTTCATGGGTGGGCTTTCACCCTCAAACAATTCTCTGAGATGTATGCTGACAAATTCAAGATTGACCTTGTCAAGCTTATGAACAGGTTATGGGGAGAAAACTTTTTCAACCCTCAAACGAAGAAGTGGTCAAAACAAAAGGATGATGACAACAAACGTTCATTTTGCATGTACGTTTTGGATCCTATCTACAAGGTGTTCGATGCCATCATGAAATTTAAGAAAGAGGAGATTGATGATCTTCTTAAGAAGATTGGAGTCACAATCAAGCATGAGGATTCCGACAAAGATGGCAAAGCTTTGCTGAAGGTTGTGATGCGCTCTTGGTTGCCTGCTGGTGAAGCTCTGCTTCAGATGATTGCCATTCATTTACCATCACCTGTAGTGGCCCAGAAATATCGTATGGAGATGTTATATGAGGGACCCCACGATGATGAAGCTGCCATTGGTATCAAGAGCTGTGATCCTGAAGCCCCACTGATGATGTACGTGAGCAAGATGGTGCCGACCTCCGACAAAGGTCGTTTCTACGCCTTTGGACGCGTTTTCTCTGGCAAGGTTGTTACCGGACAAAAAGCTCGCATCATGGGACCAAACTTTACACCTGGAAAGAAAGAGGACTTGTATGAGAAGACTATCCAGCGTACAATCCTTATGATGGGACGTTATGTTGAAGCTATTGAGGATGTGCCCTCTGGTAACATCTGTGGTCTTGTTGGAGTCGATCAGTTCTTAGTCAAGACTGGTACCATCACCACTTTCAAGAATGCCCACAACATGAAGGTGATGAAATTCAGTGTATCACCAGTCGTGCGTGTCGCTGTTGAGCCCAAAAACCCTGCTGATCTGCCCAAGCTAGTAGAAGGTCTTAAACGTCTGGCTAAATCTGACCCCATGGTGCAGTGTATTAATGAAGAATCAGGAGAACACATTGTCGCTGGTGCTGGAGAACTCCATCTTGAGATCTGTCTTAAGGATCTTGAGGAGGACCATGCTTGCATTCCAATCAAGAAGTCTGACCCTGTCGTGTCGTACCGTGAGACCGTAGCTGAGGAATCGGACCAGCTCTGTCTCTCAAAGTCGCCCAACAAGCACAACCGTCTGTTCATGAAGGCTCAGCCCATGCCTGATGGTCTGCCAGAGGACATTGATGAGGGTCGCGTGAATCCCCGCGATGACTTCAAGACTCGCGCTCGGTATCTTACAGAAAAGTACGAATATGATGTTACCGAAGCCCGTAAGATTTGGTGCTTTGGCCCCGAGGGTACCGGCCCCAACATCCTGGTGGATTGCTCCAAAGGAGTTCAGTACCTCAATGAAATTAAGGACTCTGTTGTGGCTGGATTCCAGTGGGCCGCTAAGGAAGGAGTTATGGCTGAAGAGAATTTGCGTGGTGTTAGATTCAACATCTATGATGTAACACTCCATACTGATGCCATCCATAGAGGTGGTGGCCAAATCATTCCAACAACTAGAAGATGCTTGTACGCATGTCTGCTAACTGCTCAGCCCCGTCTTATGGAGCCTGTATATCTTTGTGAAATTCAGTGTCCTGAAGTAGCTGTGGGTGGTATCTACGGTGTACTGAACAGACGTCGTGGTCACGTTTTCGAAGAGTCCCAGGTGGCAGGTACACCTATGTTCATTGTGAAGGCCTACCTACCTGTCAATGAGTCGTTCGGTTTTACTGCCGATTTGCGTTCCAACACCGGCGGACAGGCCTTCCCGCAGTGCGTATTCGACCATTGGCAGGTCCTCCCTGGAGACCCGTGCGAACCTCAGAGCAAGCCCTACAACGTTGTACAGGAAACGAGAAAGAGGAAAGGATTGAAGGAAGGTCTCCCAGACTTAACTCAATATTTGGACAAATTGTAA
Protein
MVNFTVDEIRGMMDKKRNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAGARAGETRFTDTRKDEQDRCITIKSTAISMFFELEEKDLVFITNPDQREKSEKGFLINLIDSPGHVDFSSEVTAALRVTDGALVVVDCVSGVCVQTETVLRQAIAERIKPILFMNKMDRALLELQLEAEELYQTFQRIVENVNVIIATYNDDGGPMGEVRVDPSKGSVGFGSGLHGWAFTLKQFSEMYADKFKIDLVKLMNRLWGENFFNPQTKKWSKQKDDDNKRSFCMYVLDPIYKVFDAIMKFKKEEIDDLLKKIGVTIKHEDSDKDGKALLKVVMRSWLPAGEALLQMIAIHLPSPVVAQKYRMEMLYEGPHDDEAAIGIKSCDPEAPLMMYVSKMVPTSDKGRFYAFGRVFSGKVVTGQKARIMGPNFTPGKKEDLYEKTIQRTILMMGRYVEAIEDVPSGNICGLVGVDQFLVKTGTITTFKNAHNMKVMKFSVSPVVRVAVEPKNPADLPKLVEGLKRLAKSDPMVQCINEESGEHIVAGAGELHLEICLKDLEEDHACIPIKKSDPVVSYRETVAEESDQLCLSKSPNKHNRLFMKAQPMPDGLPEDIDEGRVNPRDDFKTRARYLTEKYEYDVTEARKIWCFGPEGTGPNILVDCSKGVQYLNEIKDSVVAGFQWAAKEGVMAEENLRGVRFNIYDVTLHTDAIHRGGGQIIPTTRRCLYACLLTAQPRLMEPVYLCEIQCPEVAVGGIYGVLNRRRGHVFEESQVAGTPMFIVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQVLPGDPCEPQSKPYNVVQETRKRKGLKEGLPDLTQYLDKL
Summary
Description
Catalyzes the GTP-dependent ribosomal translocation step during translation elongation. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the newly formed A-site-bound peptidyl-tRNA and P-site-bound deacylated tRNA move to the P and E sites, respectively. Catalyzes the coordinated movement of the two tRNA molecules, the mRNA and conformational changes in the ribosome (By similarity).
Similarity
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. EF-G/EF-2 subfamily.
Keywords
Complete proteome
Cytoplasm
Direct protein sequencing
Elongation factor
GTP-binding
Nucleotide-binding
Phosphoprotein
Protein biosynthesis
Reference proteome
3D-structure
Feature
chain Translation elongation factor 2
Uniprot
F8SKB6
Q1HPK6
H9J3S8
A0A3G1T184
A0A2A4K5I8
Q86M26
+ More
A0A3S2LAA0 I4DIN3 A0A194PNI0 I4DM49 A0A2A4K5U4 A0A2W1BBG8 A0A194QYQ6 A0A0C9RJ98 V5GJC6 A0A1B0CCP0 B4IIG4 A0A1W4VEY1 G9C5D4 B4KF22 A0A0N0BKR4 A0A1Y1MPS5 P13060 U5EVG5 B4Q4F6 B4P6G4 K7J0G8 A0A2J7RKQ6 B4LUQ2 A0A2A3EIV9 A0A088A5X7 V9IF19 A0A0U3SX64 B3NKS1 A0A0M3QTX3 A0A1L8DS92 A0A1B6M4W8 A0A067QUC2 A0A1B0BN33 A0A348G614 B4JB96 A0A1A9ZFD3 D3TP87 A0A1A9XQQ0 A0A1A9VF37 D6WRR0 B3ML86 A0A0J7KV60 A0A0A1XQE3 A0A1B6FPH2 A0A3B0JGU4 A0A034VQU3 E2C8M6 A0A310SX29 A0A1L8EET8 A0A1L8EHV8 A0A170Y4A6 A0A023FB32 A0A1B6DMG0 A0A069DWL1 R4FQU1 A0A336M255 A0A1L8EHS5 B4N128 A0A158NI82 F4X3C2 B5DK66 B4GKX8 A0A1L8EF33 A0A3L8D8X6 N6SVM6 A0A0L0C5C9 T1PHN3 A0A182GYZ5 A0A1I8PSD9 A0A154P3Q8 R4WCQ3 A0A1D1Z8P1 W8C9B5 A0A0K8TQ27 Q0IFN2 Q95P39 A0A0A9W869 E9JB57 Q9BME7 A0A151WR61 A0A182H5X9 A0A1Y9H9K7 A0A1Q3FDD7 A0A182F5A8 A0A1Q3FDD6 A0A1Q3FDC3 A0A182NEL2 A0A2M3ZY68 W5J5L8 Q8T4R9 A0A2M3YX25 B0W238 Q8T4S0 T1DJH7
A0A3S2LAA0 I4DIN3 A0A194PNI0 I4DM49 A0A2A4K5U4 A0A2W1BBG8 A0A194QYQ6 A0A0C9RJ98 V5GJC6 A0A1B0CCP0 B4IIG4 A0A1W4VEY1 G9C5D4 B4KF22 A0A0N0BKR4 A0A1Y1MPS5 P13060 U5EVG5 B4Q4F6 B4P6G4 K7J0G8 A0A2J7RKQ6 B4LUQ2 A0A2A3EIV9 A0A088A5X7 V9IF19 A0A0U3SX64 B3NKS1 A0A0M3QTX3 A0A1L8DS92 A0A1B6M4W8 A0A067QUC2 A0A1B0BN33 A0A348G614 B4JB96 A0A1A9ZFD3 D3TP87 A0A1A9XQQ0 A0A1A9VF37 D6WRR0 B3ML86 A0A0J7KV60 A0A0A1XQE3 A0A1B6FPH2 A0A3B0JGU4 A0A034VQU3 E2C8M6 A0A310SX29 A0A1L8EET8 A0A1L8EHV8 A0A170Y4A6 A0A023FB32 A0A1B6DMG0 A0A069DWL1 R4FQU1 A0A336M255 A0A1L8EHS5 B4N128 A0A158NI82 F4X3C2 B5DK66 B4GKX8 A0A1L8EF33 A0A3L8D8X6 N6SVM6 A0A0L0C5C9 T1PHN3 A0A182GYZ5 A0A1I8PSD9 A0A154P3Q8 R4WCQ3 A0A1D1Z8P1 W8C9B5 A0A0K8TQ27 Q0IFN2 Q95P39 A0A0A9W869 E9JB57 Q9BME7 A0A151WR61 A0A182H5X9 A0A1Y9H9K7 A0A1Q3FDD7 A0A182F5A8 A0A1Q3FDD6 A0A1Q3FDC3 A0A182NEL2 A0A2M3ZY68 W5J5L8 Q8T4R9 A0A2M3YX25 B0W238 Q8T4S0 T1DJH7
Pubmed
11280994
19121390
12542638
22651552
26354079
28756777
+ More
17994087 21835214 28004739 2508059 10731132 12537572 12537569 23636399 22936249 17550304 20075255 26694822 24845553 20353571 18362917 19820115 25830018 25348373 20798317 25474469 26334808 21347285 21719571 15632085 30249741 23537049 26108605 25315136 26483478 23691247 24495485 26369729 17510324 25401762 26823975 21282665 20920257 23761445
17994087 21835214 28004739 2508059 10731132 12537572 12537569 23636399 22936249 17550304 20075255 26694822 24845553 20353571 18362917 19820115 25830018 25348373 20798317 25474469 26334808 21347285 21719571 15632085 30249741 23537049 26108605 25315136 26483478 23691247 24495485 26369729 17510324 25401762 26823975 21282665 20920257 23761445
EMBL
HQ638206
AEH16569.1
DQ515926
DQ443396
ABF51485.1
ABF71565.1
+ More
BABH01028837 MG846894 AXY94746.1 NWSH01000149 PCG79043.1 AY078407 AAL83698.1 RSAL01000070 RVE49182.1 AK401151 BAM17773.1 KQ459598 KPI94543.1 AK402367 BAM18989.1 PCG79042.1 KZ150162 PZC72689.1 KQ460949 KPJ10424.1 GBYB01008285 GBYB01014149 JAG78052.1 JAG83916.1 GALX01004272 JAB64194.1 AJWK01006930 CH480843 EDW49714.1 HQ851375 AEV89753.1 CH933807 EDW11991.1 KQ435697 KOX80791.1 GEZM01024973 JAV87662.1 X15805 AE014134 AY075481 GANO01001036 JAB58835.1 CM000361 CM002910 EDX05753.1 KMY91369.1 CM000158 EDW89921.1 NEVH01002717 PNF41379.1 CH940649 EDW63220.1 KZ288229 PBC31657.1 JR040732 JR040733 AEY59267.1 AEY59268.1 KU218666 ALX00046.1 CH954179 EDV54375.1 CP012523 ALC39663.1 GFDF01004775 JAV09309.1 GEBQ01009006 JAT30971.1 KK853253 KDR09305.1 JXJN01017195 FX985553 BBF97887.1 CH916368 EDW02901.1 CCAG010016184 EZ423239 ADD19515.1 KQ971354 EFA05965.1 CH902620 EDV31704.1 LBMM01002919 KMQ94183.1 GBXI01001192 JAD13100.1 GECZ01017671 JAS52098.1 OUUW01000006 SPP81577.1 GAKP01015014 JAC43938.1 GL453666 EFN75756.1 KQ759828 OAD62656.1 GFDG01001635 JAV17164.1 GFDG01000565 JAV18234.1 GEMB01003680 JAR99539.1 GBBI01000579 JAC18133.1 GEDC01010435 JAS26863.1 GBGD01000461 JAC88428.1 GAHY01000224 JAA77286.1 UFQT01000346 SSX23431.1 GFDG01000568 JAV18231.1 CH963920 EDW78190.1 ADTU01016076 GL888609 EGI59103.1 CH379062 EDY70690.2 CH479184 EDW37294.1 GFDG01001600 JAV17199.1 QOIP01000012 RLU16339.1 APGK01054910 APGK01054911 KB741253 KB632340 ENN71809.1 ERL92951.1 JRES01000973 KNC26644.1 KA647398 AFP62027.1 JXUM01098525 JXUM01098526 JXUM01098527 KQ564427 KXJ72294.1 KQ434809 KZC06579.1 AK417058 BAN20273.1 GDJX01004676 JAT63260.1 GAMC01002304 JAC04252.1 GDAI01001362 JAI16241.1 CH477306 EAT44110.1 EAT44111.1 AY040342 AAK77225.1 GBHO01040921 GDHC01016664 JAG02683.1 JAQ01965.1 GL770938 EFZ09897.1 AF331798 AAK01430.1 KQ982813 KYQ50291.1 JXUM01112929 KQ565638 KXJ70824.1 AXCN02000850 GFDL01009460 JAV25585.1 GFDL01009459 JAV25586.1 GFDL01009464 JAV25581.1 GGFK01000091 MBW33412.1 ADMH02002093 ETN59286.1 AY064104 AAL85605.1 GGFM01000070 MBW20821.1 DS231825 EDS28166.1 AY064103 AAL85604.1 GAMD01001436 JAB00155.1
BABH01028837 MG846894 AXY94746.1 NWSH01000149 PCG79043.1 AY078407 AAL83698.1 RSAL01000070 RVE49182.1 AK401151 BAM17773.1 KQ459598 KPI94543.1 AK402367 BAM18989.1 PCG79042.1 KZ150162 PZC72689.1 KQ460949 KPJ10424.1 GBYB01008285 GBYB01014149 JAG78052.1 JAG83916.1 GALX01004272 JAB64194.1 AJWK01006930 CH480843 EDW49714.1 HQ851375 AEV89753.1 CH933807 EDW11991.1 KQ435697 KOX80791.1 GEZM01024973 JAV87662.1 X15805 AE014134 AY075481 GANO01001036 JAB58835.1 CM000361 CM002910 EDX05753.1 KMY91369.1 CM000158 EDW89921.1 NEVH01002717 PNF41379.1 CH940649 EDW63220.1 KZ288229 PBC31657.1 JR040732 JR040733 AEY59267.1 AEY59268.1 KU218666 ALX00046.1 CH954179 EDV54375.1 CP012523 ALC39663.1 GFDF01004775 JAV09309.1 GEBQ01009006 JAT30971.1 KK853253 KDR09305.1 JXJN01017195 FX985553 BBF97887.1 CH916368 EDW02901.1 CCAG010016184 EZ423239 ADD19515.1 KQ971354 EFA05965.1 CH902620 EDV31704.1 LBMM01002919 KMQ94183.1 GBXI01001192 JAD13100.1 GECZ01017671 JAS52098.1 OUUW01000006 SPP81577.1 GAKP01015014 JAC43938.1 GL453666 EFN75756.1 KQ759828 OAD62656.1 GFDG01001635 JAV17164.1 GFDG01000565 JAV18234.1 GEMB01003680 JAR99539.1 GBBI01000579 JAC18133.1 GEDC01010435 JAS26863.1 GBGD01000461 JAC88428.1 GAHY01000224 JAA77286.1 UFQT01000346 SSX23431.1 GFDG01000568 JAV18231.1 CH963920 EDW78190.1 ADTU01016076 GL888609 EGI59103.1 CH379062 EDY70690.2 CH479184 EDW37294.1 GFDG01001600 JAV17199.1 QOIP01000012 RLU16339.1 APGK01054910 APGK01054911 KB741253 KB632340 ENN71809.1 ERL92951.1 JRES01000973 KNC26644.1 KA647398 AFP62027.1 JXUM01098525 JXUM01098526 JXUM01098527 KQ564427 KXJ72294.1 KQ434809 KZC06579.1 AK417058 BAN20273.1 GDJX01004676 JAT63260.1 GAMC01002304 JAC04252.1 GDAI01001362 JAI16241.1 CH477306 EAT44110.1 EAT44111.1 AY040342 AAK77225.1 GBHO01040921 GDHC01016664 JAG02683.1 JAQ01965.1 GL770938 EFZ09897.1 AF331798 AAK01430.1 KQ982813 KYQ50291.1 JXUM01112929 KQ565638 KXJ70824.1 AXCN02000850 GFDL01009460 JAV25585.1 GFDL01009459 JAV25586.1 GFDL01009464 JAV25581.1 GGFK01000091 MBW33412.1 ADMH02002093 ETN59286.1 AY064104 AAL85605.1 GGFM01000070 MBW20821.1 DS231825 EDS28166.1 AY064103 AAL85604.1 GAMD01001436 JAB00155.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000092461
+ More
UP000001292 UP000192221 UP000009192 UP000053105 UP000000803 UP000000304 UP000002282 UP000002358 UP000235965 UP000008792 UP000242457 UP000005203 UP000008711 UP000092553 UP000027135 UP000092460 UP000001070 UP000092445 UP000092444 UP000092443 UP000078200 UP000007266 UP000007801 UP000036403 UP000268350 UP000008237 UP000007798 UP000005205 UP000007755 UP000001819 UP000008744 UP000279307 UP000019118 UP000030742 UP000037069 UP000095301 UP000069940 UP000249989 UP000095300 UP000076502 UP000008820 UP000075809 UP000075886 UP000069272 UP000075884 UP000000673 UP000002320
UP000001292 UP000192221 UP000009192 UP000053105 UP000000803 UP000000304 UP000002282 UP000002358 UP000235965 UP000008792 UP000242457 UP000005203 UP000008711 UP000092553 UP000027135 UP000092460 UP000001070 UP000092445 UP000092444 UP000092443 UP000078200 UP000007266 UP000007801 UP000036403 UP000268350 UP000008237 UP000007798 UP000005205 UP000007755 UP000001819 UP000008744 UP000279307 UP000019118 UP000030742 UP000037069 UP000095301 UP000069940 UP000249989 UP000095300 UP000076502 UP000008820 UP000075809 UP000075886 UP000069272 UP000075884 UP000000673 UP000002320
PRIDE
Interpro
IPR027417
P-loop_NTPase
+ More
IPR035647 EFG_III/V
IPR020568 Ribosomal_S5_D2-typ_fold
IPR005517 Transl_elong_EFG/EF2_IV
IPR041095 EFG_II
IPR004161 EFTu-like_2
IPR009000 Transl_B-barrel_sf
IPR005225 Small_GTP-bd_dom
IPR000640 EFG_V-like
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR000795 TF_GTP-bd_dom
IPR031157 G_TR_CS
IPR035647 EFG_III/V
IPR020568 Ribosomal_S5_D2-typ_fold
IPR005517 Transl_elong_EFG/EF2_IV
IPR041095 EFG_II
IPR004161 EFTu-like_2
IPR009000 Transl_B-barrel_sf
IPR005225 Small_GTP-bd_dom
IPR000640 EFG_V-like
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR000795 TF_GTP-bd_dom
IPR031157 G_TR_CS
Gene 3D
ProteinModelPortal
F8SKB6
Q1HPK6
H9J3S8
A0A3G1T184
A0A2A4K5I8
Q86M26
+ More
A0A3S2LAA0 I4DIN3 A0A194PNI0 I4DM49 A0A2A4K5U4 A0A2W1BBG8 A0A194QYQ6 A0A0C9RJ98 V5GJC6 A0A1B0CCP0 B4IIG4 A0A1W4VEY1 G9C5D4 B4KF22 A0A0N0BKR4 A0A1Y1MPS5 P13060 U5EVG5 B4Q4F6 B4P6G4 K7J0G8 A0A2J7RKQ6 B4LUQ2 A0A2A3EIV9 A0A088A5X7 V9IF19 A0A0U3SX64 B3NKS1 A0A0M3QTX3 A0A1L8DS92 A0A1B6M4W8 A0A067QUC2 A0A1B0BN33 A0A348G614 B4JB96 A0A1A9ZFD3 D3TP87 A0A1A9XQQ0 A0A1A9VF37 D6WRR0 B3ML86 A0A0J7KV60 A0A0A1XQE3 A0A1B6FPH2 A0A3B0JGU4 A0A034VQU3 E2C8M6 A0A310SX29 A0A1L8EET8 A0A1L8EHV8 A0A170Y4A6 A0A023FB32 A0A1B6DMG0 A0A069DWL1 R4FQU1 A0A336M255 A0A1L8EHS5 B4N128 A0A158NI82 F4X3C2 B5DK66 B4GKX8 A0A1L8EF33 A0A3L8D8X6 N6SVM6 A0A0L0C5C9 T1PHN3 A0A182GYZ5 A0A1I8PSD9 A0A154P3Q8 R4WCQ3 A0A1D1Z8P1 W8C9B5 A0A0K8TQ27 Q0IFN2 Q95P39 A0A0A9W869 E9JB57 Q9BME7 A0A151WR61 A0A182H5X9 A0A1Y9H9K7 A0A1Q3FDD7 A0A182F5A8 A0A1Q3FDD6 A0A1Q3FDC3 A0A182NEL2 A0A2M3ZY68 W5J5L8 Q8T4R9 A0A2M3YX25 B0W238 Q8T4S0 T1DJH7
A0A3S2LAA0 I4DIN3 A0A194PNI0 I4DM49 A0A2A4K5U4 A0A2W1BBG8 A0A194QYQ6 A0A0C9RJ98 V5GJC6 A0A1B0CCP0 B4IIG4 A0A1W4VEY1 G9C5D4 B4KF22 A0A0N0BKR4 A0A1Y1MPS5 P13060 U5EVG5 B4Q4F6 B4P6G4 K7J0G8 A0A2J7RKQ6 B4LUQ2 A0A2A3EIV9 A0A088A5X7 V9IF19 A0A0U3SX64 B3NKS1 A0A0M3QTX3 A0A1L8DS92 A0A1B6M4W8 A0A067QUC2 A0A1B0BN33 A0A348G614 B4JB96 A0A1A9ZFD3 D3TP87 A0A1A9XQQ0 A0A1A9VF37 D6WRR0 B3ML86 A0A0J7KV60 A0A0A1XQE3 A0A1B6FPH2 A0A3B0JGU4 A0A034VQU3 E2C8M6 A0A310SX29 A0A1L8EET8 A0A1L8EHV8 A0A170Y4A6 A0A023FB32 A0A1B6DMG0 A0A069DWL1 R4FQU1 A0A336M255 A0A1L8EHS5 B4N128 A0A158NI82 F4X3C2 B5DK66 B4GKX8 A0A1L8EF33 A0A3L8D8X6 N6SVM6 A0A0L0C5C9 T1PHN3 A0A182GYZ5 A0A1I8PSD9 A0A154P3Q8 R4WCQ3 A0A1D1Z8P1 W8C9B5 A0A0K8TQ27 Q0IFN2 Q95P39 A0A0A9W869 E9JB57 Q9BME7 A0A151WR61 A0A182H5X9 A0A1Y9H9K7 A0A1Q3FDD7 A0A182F5A8 A0A1Q3FDD6 A0A1Q3FDC3 A0A182NEL2 A0A2M3ZY68 W5J5L8 Q8T4R9 A0A2M3YX25 B0W238 Q8T4S0 T1DJH7
PDB
4V6W
E-value=0,
Score=4092
Ontologies
GO
Topology
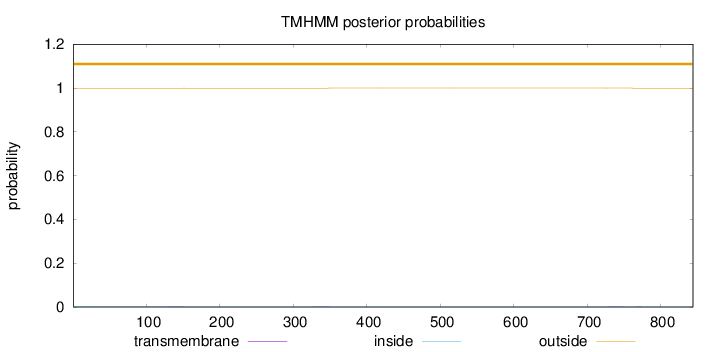
Subcellular location
Cytoplasm
Length:
844
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02486
Exp number, first 60 AAs:
0.00083
Total prob of N-in:
0.00082
outside
1 - 844
Population Genetic Test Statistics
Pi
23.246398
Theta
21.731521
Tajima's D
0.19095
CLR
1.023891
CSRT
0.431578421078946
Interpretation
Uncertain
