Gene
KWMTBOMO11729
Annotation
hypothetical_protein_RF55_12429_[Lasius_niger]
Location in the cell
Mitochondrial Reliability : 2.132
Sequence
CDS
ATGGAATCGTACCAATTACATGGTTTTGCTGACAGTTCGGAGGCTGGCTATGGAGCTGTGGTTTATCTTCGAACTCTTAATTCCGATGGTCATATAGGAGTATCCTTTTTATGTGCTAAGTCTCGCGTGGCCCCAATTAGGAGAGTTTCATTGCCGAGACTAGAGCTCTGCGCTGCGGTTCTGCTCGCTGACCTGCTCAAATTCGTAAAAGAAGTGTATGTACCGTTACTTCCGCTTTCTGATACATTTGCTTGGTCAGATTCAACGGTAACTCTGTCATGGATTCGTGGCCATTCCTCGCGTTGGAAAACATTTGTTGCTAATAGGGTTAGTCATATCCAAGAAACTGTCCCCGAAACTCACTGGAATCACGTGCGCTCTAATGAAAATCCTGCGGACATAGCTTCCCGCGGTACGTGTCCGAGCGACCTACTCCAGAGTGAGTTATGGTGGGCGGGACCGAGGTGGCTTCGGCAACCTTCATCTTCATGGCCTTCAAATTCCGGTCTCGAGGTGAACGAGACTGATATGCTTGCCGAGGAACGCTGTTTTTCGTTGGTCGTGAATGATGAACCTAACTTTATCGATTCTTTGCTCGAAAGGTTTTCATCAATTCGTAAGGTTCAACGTATTATTGGTTATTGTCTTCGTTTTATCAGAAATGCTAACAAAAAACAGACACGTGTTCCGGGTACCTTTTTAGGTCGTGCAGAACTTCATGGGGCCCTGATGACGATTACCAAGCGAATTCAATTTCTCAACTTTCAGGATGAAATTCGTAAGCTCGAGAATGGTATACCATTACCCAAACCGTGGAGAAAACTCAATCCATTCCTGGACGACCAAGGAGTTGTCAGGGTGGGCGGCCGTTTGTCTCGTTCTGGTCTTGAGTTTAGTCATAAGCATCCTGCTCTGTTGCCGCGTCATTCTAGGTTCACATACATGATCATCGAAGCAATTCACAAAGAGAATTGTCATCCTGGGTTGGGAACGACGCAATATCTTTTGATGCAGCTGTTTTGGGTATTCTCTTCTAAACGTGCTATTCGTCATTGCTTGTCAAAATGTGTTTCGTGTTATAAAACTAATCCGAAATTATTGGAACCATTCATGAGTGACTTGCCGGCTTCTCGTGTGAATCAAGTCAAACCATTCTCTATTGTTGGCGTGGACTACGCAGGTCCATTCCAAATCAAACTCGGTAAGCATCGCGGTGCCAAAATCGCTCGTGTCTCCAAGCTTCACGCCAACCCTGATGGCGTGGTTCGTATTGCCACTGTCCGTACAGCCAATGGCAAATTCTACACTAGGCCATTAACGAAGTTAGCTCCCCTACCTATTTATTCTGATTAA
Protein
MESYQLHGFADSSEAGYGAVVYLRTLNSDGHIGVSFLCAKSRVAPIRRVSLPRLELCAAVLLADLLKFVKEVYVPLLPLSDTFAWSDSTVTLSWIRGHSSRWKTFVANRVSHIQETVPETHWNHVRSNENPADIASRGTCPSDLLQSELWWAGPRWLRQPSSSWPSNSGLEVNETDMLAEERCFSLVVNDEPNFIDSLLERFSSIRKVQRIIGYCLRFIRNANKKQTRVPGTFLGRAELHGALMTITKRIQFLNFQDEIRKLENGIPLPKPWRKLNPFLDDQGVVRVGGRLSRSGLEFSHKHPALLPRHSRFTYMIIEAIHKENCHPGLGTTQYLLMQLFWVFSSKRAIRHCLSKCVSCYKTNPKLLEPFMSDLPASRVNQVKPFSIVGVDYAGPFQIKLGKHRGAKIARVSKLHANPDGVVRIATVRTANGKFYTRPLTKLAPLPIYSD
Summary
Uniprot
V5GHM7
A0A0J7KD57
V5GC07
A0A1Y1KMX0
A0A2S2R5S9
A0A0J7K7U0
+ More
A0A0J7KC70 D6WY83 A0A0J7KDN6 A0A1B6MFR0 A0A1Y1LIG5 A0A146M0N4 E2ARC4 A0A1B6JE15 A0A023F096 J9JNP4 A0A2S2PED9 A0A1B6GMH9 A0A2S2QPS5 X1WY95 X1XIA7 X1XU26 X1XMM8 A0A2S2N9D0 J9LPA9 X1WSS7 X1XTX7 A0A2S2PM34 J9KLD6 A0A3L8DZP4 A0A1Y1KVX6 E1ZWH0 A0A1B6MUZ9 A0A1B6KZV2 J9KD60 A0A0J7KBV8 A0A182IER0 A0A0J7K7S0 A0A0J7KCV7 A0A0J7KBZ0 A0A0J7N189 W8BHD4 J9JLG6 A0A0J7K9A7 A0A1Y1L2Y2 A0A3L8D7N7 A0A0J7K907 A0A1S3T8T7 A0A0J7KBS8 A0A2S2NLW5 A0A0J7KCR4 A0A226D417 A0A3L8D7Q9 A0A1Y1LBZ3 X1XEA7 A0A0J7K952 A0A1B6CM38 A0A1B6LDH0 A0A2S2PUJ2 A0A226CW75 A0A182RKT7 A0A0J7KJY9 A0A0J7JZS0 A0A0J7KD75 A0A226CWA5 J9JVD6 A0A226DMG9 A0A0J7N5C9 A0A0J7N536 A0A0J7N2B8 A0A226E226 A0A0A9VS37 A0A182HA33 A0A1S3DNE5 X1WYQ8 A0A2S2P3E6 A0A1Y1N4T1 A0A0A9WIR2 A0A0V0VDM6 A0A0J7KCI2 A0A0J7N465 A0A1U8N896 A0A2S2NCY5 X1XU46 A0A0J7K9I5 W8B1J3 A0A0J7KIZ4 J9KCB6 X1WNS5 K7J7V9 A0A0J7KNK6 A0A2L2YSY1 A0A0V0UWT4 A0A226EUF5 A0A1Y1N1H8 D7EKN1 A0A1S3DIZ2 A0A087T9T6 A0A1Y1MTT1 A0A0N0PAG9
A0A0J7KC70 D6WY83 A0A0J7KDN6 A0A1B6MFR0 A0A1Y1LIG5 A0A146M0N4 E2ARC4 A0A1B6JE15 A0A023F096 J9JNP4 A0A2S2PED9 A0A1B6GMH9 A0A2S2QPS5 X1WY95 X1XIA7 X1XU26 X1XMM8 A0A2S2N9D0 J9LPA9 X1WSS7 X1XTX7 A0A2S2PM34 J9KLD6 A0A3L8DZP4 A0A1Y1KVX6 E1ZWH0 A0A1B6MUZ9 A0A1B6KZV2 J9KD60 A0A0J7KBV8 A0A182IER0 A0A0J7K7S0 A0A0J7KCV7 A0A0J7KBZ0 A0A0J7N189 W8BHD4 J9JLG6 A0A0J7K9A7 A0A1Y1L2Y2 A0A3L8D7N7 A0A0J7K907 A0A1S3T8T7 A0A0J7KBS8 A0A2S2NLW5 A0A0J7KCR4 A0A226D417 A0A3L8D7Q9 A0A1Y1LBZ3 X1XEA7 A0A0J7K952 A0A1B6CM38 A0A1B6LDH0 A0A2S2PUJ2 A0A226CW75 A0A182RKT7 A0A0J7KJY9 A0A0J7JZS0 A0A0J7KD75 A0A226CWA5 J9JVD6 A0A226DMG9 A0A0J7N5C9 A0A0J7N536 A0A0J7N2B8 A0A226E226 A0A0A9VS37 A0A182HA33 A0A1S3DNE5 X1WYQ8 A0A2S2P3E6 A0A1Y1N4T1 A0A0A9WIR2 A0A0V0VDM6 A0A0J7KCI2 A0A0J7N465 A0A1U8N896 A0A2S2NCY5 X1XU46 A0A0J7K9I5 W8B1J3 A0A0J7KIZ4 J9KCB6 X1WNS5 K7J7V9 A0A0J7KNK6 A0A2L2YSY1 A0A0V0UWT4 A0A226EUF5 A0A1Y1N1H8 D7EKN1 A0A1S3DIZ2 A0A087T9T6 A0A1Y1MTT1 A0A0N0PAG9
Pubmed
EMBL
GALX01004947
JAB63519.1
LBMM01009442
KMQ88136.1
GALX01000776
JAB67690.1
+ More
GEZM01085145 GEZM01085143 JAV60187.1 GGMS01016125 MBY85328.1 LBMM01012238 KMQ86334.1 LBMM01009863 KMQ87831.1 KQ971357 EFA07702.1 LBMM01009202 KMQ88311.1 GEBQ01005223 JAT34754.1 GEZM01054765 JAV73413.1 GDHC01006469 JAQ12160.1 GL442026 EFN64015.1 GECU01010337 JAS97369.1 GBBI01004276 JAC14436.1 ABLF02005684 GGMR01015135 MBY27754.1 GECZ01006169 JAS63600.1 GGMS01010524 MBY79727.1 ABLF02030854 ABLF02067347 ABLF02035110 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 ABLF02017373 GGMR01000737 MBY13356.1 ABLF02035780 ABLF02017056 ABLF02006304 ABLF02036268 GGMR01017890 MBY30509.1 ABLF02013940 ABLF02036313 ABLF02066938 ABLF02066950 QOIP01000002 RLU25950.1 GEZM01072348 JAV65549.1 GL434835 EFN74469.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 ABLF02041863 LBMM01009941 KMQ87772.1 APCN01009161 LBMM01012392 KMQ86241.1 LBMM01009403 KMQ88162.1 LBMM01009964 KMQ87756.1 LBMM01012077 KMQ86420.1 GAMC01005870 JAC00686.1 ABLF02030698 ABLF02030705 ABLF02030709 LBMM01011283 KMQ86917.1 GEZM01068861 GEZM01068860 JAV66730.1 QOIP01000012 RLU16284.1 LBMM01011516 KMQ86784.1 LBMM01009888 KMQ87813.1 GGMR01005535 MBY18154.1 LBMM01009551 KMQ88039.1 LNIX01000036 OXA39969.1 RLU15928.1 GEZM01060254 JAV71154.1 ABLF02006652 ABLF02006653 ABLF02026952 ABLF02038350 ABLF02038362 LBMM01011245 KMQ86943.1 GEDC01022769 JAS14529.1 GEBQ01018232 JAT21745.1 GGMR01020500 MBY33119.1 LNIX01000063 OXA37199.1 LBMM01006490 KMQ90572.1 LBMM01018894 KMQ83668.1 LBMM01009212 KMQ88302.1 LNIX01000069 OXA36894.1 ABLF02008594 LNIX01000016 OXA46198.1 LBMM01009746 KMQ87900.1 LBMM01009898 KMQ87805.1 LBMM01011492 KMQ86800.1 LNIX01000007 OXA51795.1 GBHO01044457 JAF99146.1 JXUM01030959 KQ560903 KXJ80414.1 ABLF02028666 ABLF02066713 GGMR01011253 MBY23872.1 GEZM01012999 JAV92824.1 GBHO01035277 JAG08327.1 JYDK01000531 KRX61644.1 LBMM01009688 KMQ87939.1 LBMM01010417 KMQ87475.1 GGMR01002421 MBY15040.1 ABLF02003339 ABLF02058001 LBMM01011244 KMQ86944.1 GAMC01011600 GAMC01011596 JAB94955.1 LBMM01006671 KMQ90403.1 ABLF02042264 ABLF02055921 AAZX01023505 LBMM01005079 KMQ91831.1 IAAA01051064 LAA11179.1 JYDK01000686 JYDK01000685 KRX55769.1 KRX55800.1 LNIX01000002 OXA60848.1 GEZM01018628 JAV90116.1 DS497760 EFA13221.1 KK114194 KFM61875.1 GEZM01021505 JAV88969.1 KQ458473 KPJ05885.1
GEZM01085145 GEZM01085143 JAV60187.1 GGMS01016125 MBY85328.1 LBMM01012238 KMQ86334.1 LBMM01009863 KMQ87831.1 KQ971357 EFA07702.1 LBMM01009202 KMQ88311.1 GEBQ01005223 JAT34754.1 GEZM01054765 JAV73413.1 GDHC01006469 JAQ12160.1 GL442026 EFN64015.1 GECU01010337 JAS97369.1 GBBI01004276 JAC14436.1 ABLF02005684 GGMR01015135 MBY27754.1 GECZ01006169 JAS63600.1 GGMS01010524 MBY79727.1 ABLF02030854 ABLF02067347 ABLF02035110 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 ABLF02017373 GGMR01000737 MBY13356.1 ABLF02035780 ABLF02017056 ABLF02006304 ABLF02036268 GGMR01017890 MBY30509.1 ABLF02013940 ABLF02036313 ABLF02066938 ABLF02066950 QOIP01000002 RLU25950.1 GEZM01072348 JAV65549.1 GL434835 EFN74469.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 ABLF02041863 LBMM01009941 KMQ87772.1 APCN01009161 LBMM01012392 KMQ86241.1 LBMM01009403 KMQ88162.1 LBMM01009964 KMQ87756.1 LBMM01012077 KMQ86420.1 GAMC01005870 JAC00686.1 ABLF02030698 ABLF02030705 ABLF02030709 LBMM01011283 KMQ86917.1 GEZM01068861 GEZM01068860 JAV66730.1 QOIP01000012 RLU16284.1 LBMM01011516 KMQ86784.1 LBMM01009888 KMQ87813.1 GGMR01005535 MBY18154.1 LBMM01009551 KMQ88039.1 LNIX01000036 OXA39969.1 RLU15928.1 GEZM01060254 JAV71154.1 ABLF02006652 ABLF02006653 ABLF02026952 ABLF02038350 ABLF02038362 LBMM01011245 KMQ86943.1 GEDC01022769 JAS14529.1 GEBQ01018232 JAT21745.1 GGMR01020500 MBY33119.1 LNIX01000063 OXA37199.1 LBMM01006490 KMQ90572.1 LBMM01018894 KMQ83668.1 LBMM01009212 KMQ88302.1 LNIX01000069 OXA36894.1 ABLF02008594 LNIX01000016 OXA46198.1 LBMM01009746 KMQ87900.1 LBMM01009898 KMQ87805.1 LBMM01011492 KMQ86800.1 LNIX01000007 OXA51795.1 GBHO01044457 JAF99146.1 JXUM01030959 KQ560903 KXJ80414.1 ABLF02028666 ABLF02066713 GGMR01011253 MBY23872.1 GEZM01012999 JAV92824.1 GBHO01035277 JAG08327.1 JYDK01000531 KRX61644.1 LBMM01009688 KMQ87939.1 LBMM01010417 KMQ87475.1 GGMR01002421 MBY15040.1 ABLF02003339 ABLF02058001 LBMM01011244 KMQ86944.1 GAMC01011600 GAMC01011596 JAB94955.1 LBMM01006671 KMQ90403.1 ABLF02042264 ABLF02055921 AAZX01023505 LBMM01005079 KMQ91831.1 IAAA01051064 LAA11179.1 JYDK01000686 JYDK01000685 KRX55769.1 KRX55800.1 LNIX01000002 OXA60848.1 GEZM01018628 JAV90116.1 DS497760 EFA13221.1 KK114194 KFM61875.1 GEZM01021505 JAV88969.1 KQ458473 KPJ05885.1
Proteomes
Pfam
Interpro
IPR041588
Integrase_H2C2
+ More
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001584 Integrase_cat-core
IPR000477 RT_dom
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR005312 DUF1759
IPR001995 Peptidase_A2_cat
IPR000727 T_SNARE_dom
IPR001969 Aspartic_peptidase_AS
IPR032403 Exo84_C
IPR016159 Cullin_repeat-like_dom_sf
IPR017907 Znf_RING_CS
IPR041373 RT_RNaseH
IPR041577 RT_RNaseH_2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001584 Integrase_cat-core
IPR000477 RT_dom
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR005312 DUF1759
IPR001995 Peptidase_A2_cat
IPR000727 T_SNARE_dom
IPR001969 Aspartic_peptidase_AS
IPR032403 Exo84_C
IPR016159 Cullin_repeat-like_dom_sf
IPR017907 Znf_RING_CS
IPR041373 RT_RNaseH
IPR041577 RT_RNaseH_2
Gene 3D
ProteinModelPortal
V5GHM7
A0A0J7KD57
V5GC07
A0A1Y1KMX0
A0A2S2R5S9
A0A0J7K7U0
+ More
A0A0J7KC70 D6WY83 A0A0J7KDN6 A0A1B6MFR0 A0A1Y1LIG5 A0A146M0N4 E2ARC4 A0A1B6JE15 A0A023F096 J9JNP4 A0A2S2PED9 A0A1B6GMH9 A0A2S2QPS5 X1WY95 X1XIA7 X1XU26 X1XMM8 A0A2S2N9D0 J9LPA9 X1WSS7 X1XTX7 A0A2S2PM34 J9KLD6 A0A3L8DZP4 A0A1Y1KVX6 E1ZWH0 A0A1B6MUZ9 A0A1B6KZV2 J9KD60 A0A0J7KBV8 A0A182IER0 A0A0J7K7S0 A0A0J7KCV7 A0A0J7KBZ0 A0A0J7N189 W8BHD4 J9JLG6 A0A0J7K9A7 A0A1Y1L2Y2 A0A3L8D7N7 A0A0J7K907 A0A1S3T8T7 A0A0J7KBS8 A0A2S2NLW5 A0A0J7KCR4 A0A226D417 A0A3L8D7Q9 A0A1Y1LBZ3 X1XEA7 A0A0J7K952 A0A1B6CM38 A0A1B6LDH0 A0A2S2PUJ2 A0A226CW75 A0A182RKT7 A0A0J7KJY9 A0A0J7JZS0 A0A0J7KD75 A0A226CWA5 J9JVD6 A0A226DMG9 A0A0J7N5C9 A0A0J7N536 A0A0J7N2B8 A0A226E226 A0A0A9VS37 A0A182HA33 A0A1S3DNE5 X1WYQ8 A0A2S2P3E6 A0A1Y1N4T1 A0A0A9WIR2 A0A0V0VDM6 A0A0J7KCI2 A0A0J7N465 A0A1U8N896 A0A2S2NCY5 X1XU46 A0A0J7K9I5 W8B1J3 A0A0J7KIZ4 J9KCB6 X1WNS5 K7J7V9 A0A0J7KNK6 A0A2L2YSY1 A0A0V0UWT4 A0A226EUF5 A0A1Y1N1H8 D7EKN1 A0A1S3DIZ2 A0A087T9T6 A0A1Y1MTT1 A0A0N0PAG9
A0A0J7KC70 D6WY83 A0A0J7KDN6 A0A1B6MFR0 A0A1Y1LIG5 A0A146M0N4 E2ARC4 A0A1B6JE15 A0A023F096 J9JNP4 A0A2S2PED9 A0A1B6GMH9 A0A2S2QPS5 X1WY95 X1XIA7 X1XU26 X1XMM8 A0A2S2N9D0 J9LPA9 X1WSS7 X1XTX7 A0A2S2PM34 J9KLD6 A0A3L8DZP4 A0A1Y1KVX6 E1ZWH0 A0A1B6MUZ9 A0A1B6KZV2 J9KD60 A0A0J7KBV8 A0A182IER0 A0A0J7K7S0 A0A0J7KCV7 A0A0J7KBZ0 A0A0J7N189 W8BHD4 J9JLG6 A0A0J7K9A7 A0A1Y1L2Y2 A0A3L8D7N7 A0A0J7K907 A0A1S3T8T7 A0A0J7KBS8 A0A2S2NLW5 A0A0J7KCR4 A0A226D417 A0A3L8D7Q9 A0A1Y1LBZ3 X1XEA7 A0A0J7K952 A0A1B6CM38 A0A1B6LDH0 A0A2S2PUJ2 A0A226CW75 A0A182RKT7 A0A0J7KJY9 A0A0J7JZS0 A0A0J7KD75 A0A226CWA5 J9JVD6 A0A226DMG9 A0A0J7N5C9 A0A0J7N536 A0A0J7N2B8 A0A226E226 A0A0A9VS37 A0A182HA33 A0A1S3DNE5 X1WYQ8 A0A2S2P3E6 A0A1Y1N4T1 A0A0A9WIR2 A0A0V0VDM6 A0A0J7KCI2 A0A0J7N465 A0A1U8N896 A0A2S2NCY5 X1XU46 A0A0J7K9I5 W8B1J3 A0A0J7KIZ4 J9KCB6 X1WNS5 K7J7V9 A0A0J7KNK6 A0A2L2YSY1 A0A0V0UWT4 A0A226EUF5 A0A1Y1N1H8 D7EKN1 A0A1S3DIZ2 A0A087T9T6 A0A1Y1MTT1 A0A0N0PAG9
Ontologies
KEGG
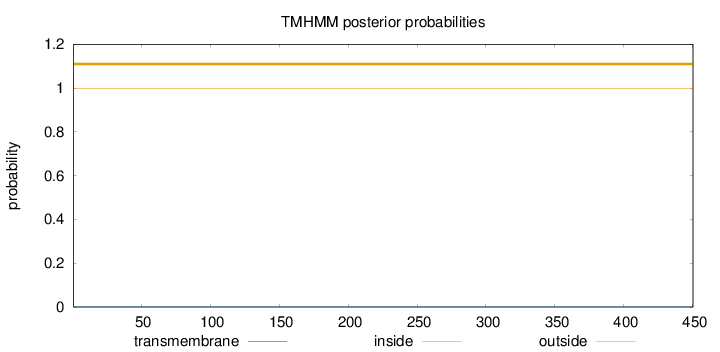
Topology
Length:
450
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03545
Exp number, first 60 AAs:
0.01425
Total prob of N-in:
0.00200
outside
1 - 450
Population Genetic Test Statistics
Pi
16.670344
Theta
26.343608
Tajima's D
-1.006293
CLR
12.539634
CSRT
0.138043097845108
Interpretation
Uncertain
