Gene
KWMTBOMO11726
Pre Gene Modal
BGIBMGA004188
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_A_member_3-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.677
Sequence
CDS
ATGAATGCAAGCAACCAAATCAAGGACGTTCTGTTAGCGGACAACCTGCACAAGAGGTACCCCAAGATCTTCGGCAAGTCCTGCAACGCCGTCAAAGGAGTCAGTTTCTCGGTTAAGAAAGGAGAATGCTTTGGCCTCCTCGGCGTGAACGGAGCCGGCAAGTCTACCACCTTCAAGATGCTGACGGCCGAGGAGTGCGCCACTCGAGGCAACATTTTCGCGAACGGCTACCACCTGAAGCGCAGCGTCGGACCGTACCTCCGCTCCCTCGGCTACTGCCCACAATTCTTCGGGCTGGACGACTTCCTGACCGGCGCCCAGAACCTGGCCTTGCTGCTGACCCTGCGCGGACTCGACGTCGTCGACGTGAACGCCGAGGTCGCGTCCTGGATACAGATCGTAGGGCTGGAGAAGTACATGCACCGGCCCGTGTCGGGCTACAGCGGCGGCTGCAAGCGGCGGCTGGCGGCGGGCGCGGCGCTGGCGGCGGGCGCGGCCGTCACGCTGCTGGACGAGCCCACCGCGGGCGTGGACGTGGCCGCGCGCCGCCGCGTGTGGGCCGCGCTGCGCCGCGCGCTGCACCGGGGACGCGCCGTCGTCATCTCCTCGCACAGGCATTAG
Protein
MNASNQIKDVLLADNLHKRYPKIFGKSCNAVKGVSFSVKKGECFGLLGVNGAGKSTTFKMLTAEECATRGNIFANGYHLKRSVGPYLRSLGYCPQFFGLDDFLTGAQNLALLLTLRGLDVVDVNAEVASWIQIVGLEKYMHRPVSGYSGGCKRRLAAGAALAAGAAVTLLDEPTAGVDVAARRRVWAALRRALHRGRAVVISSHRH
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RsmB/NOP family.
Uniprot
A0A2W1BCS5
A0A194PTT1
A0A194R3K7
A0A2A4K545
A0A0F6PQM2
A0A212F3T3
+ More
A0A2P8YJ86 A0A1B6EFI2 A0A1B6DK03 A0A1B6JS44 A0A1B6FQX5 A0A1B6FLT5 A0A1B6F8C5 A0A1B6MC35 A0A1B6G0W6 A0A1B6IZN0 A0A1B6MPR0 A0A2J7RIF1 A0A1B6CKN8 A0A2J7RIF3 A0A067QMM5 A0A1B6GZC0 A0A1B6HI75 A0A0N4XAZ4 A0A0D2WPE2 E4Y7D9 A0A1J1IJX5 A0A1W4WY30 A0A2G9V2A8 A0A0A7ARI5 E4XQ17 A0A2S2P9P8 A0A1B6GC71 A0A1Y3AWT5 A0A2S2QUV9 A0A1B6HGH5 J9K9B5 A0A1B6I525 A0A2R7W837 A0A0S7H6F6 A0A2H8TTL0 A0A0S7HAW5 A0A368GJA2 A0A0Q9XF97 A0A3P7C2C3 W6NGB2 A0A158QNT6 A0A0S7HA71 A0A0N4XMX0 A0A0C2GBZ7 A0A0D8XHI3 E4XHW7 T1IBQ4 H3C1X8 B4KIX1 H3D169 A0A0S7H654 A0A3B5PPH9 A0A3B5PRM2 A0A0L8I2K3 A0A146MPG2 M3ZRP8 A0A3Q3RQP1 A0A3B3YAD3 A0A3Q3LC94 W2T430 A0A3B3WRC0 A0A183T5C2 A0A3Q2Q6M2 A0A016WEL3 A0A016WC99 Q3YB24 A0A194PNI5 A0A183WPN0 A0A132ACH8 A0A183QDX2 A0A3P6R8D8 A0A3B3V8X0 A0A0M3K8V1 F1L0H8 A0A087XBS7 A0A3Q2YND9 A0A2B4RF03 A0A3P9HY39 A0A0K8TT98 A0A3P9L2H4 A0A3B3H814 A0A3B3I8Q3 A0A3P9L2N4 A0A3B3WR13 A0A3P9HY72 A0A3R5SPL4 A0A060YX36 A0A287AQ53 A0A161ALT0 A0A3S2MX29 A0A368GMG1 A0A1A8L276 A0A0K0E8W1
A0A2P8YJ86 A0A1B6EFI2 A0A1B6DK03 A0A1B6JS44 A0A1B6FQX5 A0A1B6FLT5 A0A1B6F8C5 A0A1B6MC35 A0A1B6G0W6 A0A1B6IZN0 A0A1B6MPR0 A0A2J7RIF1 A0A1B6CKN8 A0A2J7RIF3 A0A067QMM5 A0A1B6GZC0 A0A1B6HI75 A0A0N4XAZ4 A0A0D2WPE2 E4Y7D9 A0A1J1IJX5 A0A1W4WY30 A0A2G9V2A8 A0A0A7ARI5 E4XQ17 A0A2S2P9P8 A0A1B6GC71 A0A1Y3AWT5 A0A2S2QUV9 A0A1B6HGH5 J9K9B5 A0A1B6I525 A0A2R7W837 A0A0S7H6F6 A0A2H8TTL0 A0A0S7HAW5 A0A368GJA2 A0A0Q9XF97 A0A3P7C2C3 W6NGB2 A0A158QNT6 A0A0S7HA71 A0A0N4XMX0 A0A0C2GBZ7 A0A0D8XHI3 E4XHW7 T1IBQ4 H3C1X8 B4KIX1 H3D169 A0A0S7H654 A0A3B5PPH9 A0A3B5PRM2 A0A0L8I2K3 A0A146MPG2 M3ZRP8 A0A3Q3RQP1 A0A3B3YAD3 A0A3Q3LC94 W2T430 A0A3B3WRC0 A0A183T5C2 A0A3Q2Q6M2 A0A016WEL3 A0A016WC99 Q3YB24 A0A194PNI5 A0A183WPN0 A0A132ACH8 A0A183QDX2 A0A3P6R8D8 A0A3B3V8X0 A0A0M3K8V1 F1L0H8 A0A087XBS7 A0A3Q2YND9 A0A2B4RF03 A0A3P9HY39 A0A0K8TT98 A0A3P9L2H4 A0A3B3H814 A0A3B3I8Q3 A0A3P9L2N4 A0A3B3WR13 A0A3P9HY72 A0A3R5SPL4 A0A060YX36 A0A287AQ53 A0A161ALT0 A0A3S2MX29 A0A368GMG1 A0A1A8L276 A0A0K0E8W1
Pubmed
EMBL
KZ150130
PZC73059.1
KQ459598
KPI94545.1
KQ460949
KPJ10426.1
+ More
NWSH01000149 PCG79038.1 KM244036 AKC42329.1 AGBW02010485 OWR48406.1 PYGN01000555 PSN44315.1 GEDC01000674 JAS36624.1 GEDC01011313 JAS25985.1 GECU01005681 JAT02026.1 GECZ01017356 JAS52413.1 GECZ01018617 JAS51152.1 GECZ01023368 JAS46401.1 GEBQ01006481 JAT33496.1 GECZ01013694 JAS56075.1 GECU01015311 JAS92395.1 GEBQ01002122 JAT37855.1 NEVH01003500 PNF40614.1 GEDC01023308 JAS13990.1 PNF40610.1 KK853244 KDR09441.1 GECZ01001999 JAS67770.1 GECU01033338 JAS74368.1 UZAF01023585 VDO90494.1 KE346364 KJE92513.1 FN654307 CBY31539.1 CVRI01000048 CRK98753.1 KZ345046 PIO76516.1 KF906267 AHK05632.1 FN653099 CBY11903.1 GGMR01013564 MBY26183.1 GECZ01009744 JAS60025.1 MUJZ01056049 OTF72457.1 GGMS01012311 MBY81514.1 GECU01033938 JAS73768.1 ABLF02037149 ABLF02037152 ABLF02037161 GECU01025689 JAS82017.1 KK854443 PTY15893.1 GBYX01444795 JAO36625.1 GFXV01005779 MBW17584.1 GBYX01444793 JAO36627.1 JOJR01000128 RCN44473.1 CH933807 KRG03780.1 UYSL01006358 VDL67461.1 CAVP010058607 CDL94934.1 UZAF01017487 VDO42155.1 GBYX01444794 JAO36626.1 KN732949 KIH58590.1 KN716759 KJH41796.1 FN653053 CBY10183.1 ACPB03006481 EDW13484.2 GBYX01444792 JAO36628.1 KQ416708 KOF95609.1 GCES01164729 JAQ21593.1 KI660235 ETN76299.1 UYSU01036713 VDL98055.1 JARK01000389 EYC37453.1 EYC37454.1 DQ146416 AAZ75680.1 KPI94548.1 UZAO01078437 VDQ09963.1 JXLN01012619 KPM08691.1 UYRR01033424 VDK58736.1 JI169072 ADY43632.1 AYCK01010082 AYCK01010083 AYCK01010084 LSMT01000507 PFX16944.1 GDAI01000225 JAI17378.1 MH172491 QAA95898.1 FR916687 CDQ94049.1 AEMK02000017 KF669396 AIN44094.1 CM012445 RVE68758.1 RCN44479.1 HAEF01000612 SBR37994.1
NWSH01000149 PCG79038.1 KM244036 AKC42329.1 AGBW02010485 OWR48406.1 PYGN01000555 PSN44315.1 GEDC01000674 JAS36624.1 GEDC01011313 JAS25985.1 GECU01005681 JAT02026.1 GECZ01017356 JAS52413.1 GECZ01018617 JAS51152.1 GECZ01023368 JAS46401.1 GEBQ01006481 JAT33496.1 GECZ01013694 JAS56075.1 GECU01015311 JAS92395.1 GEBQ01002122 JAT37855.1 NEVH01003500 PNF40614.1 GEDC01023308 JAS13990.1 PNF40610.1 KK853244 KDR09441.1 GECZ01001999 JAS67770.1 GECU01033338 JAS74368.1 UZAF01023585 VDO90494.1 KE346364 KJE92513.1 FN654307 CBY31539.1 CVRI01000048 CRK98753.1 KZ345046 PIO76516.1 KF906267 AHK05632.1 FN653099 CBY11903.1 GGMR01013564 MBY26183.1 GECZ01009744 JAS60025.1 MUJZ01056049 OTF72457.1 GGMS01012311 MBY81514.1 GECU01033938 JAS73768.1 ABLF02037149 ABLF02037152 ABLF02037161 GECU01025689 JAS82017.1 KK854443 PTY15893.1 GBYX01444795 JAO36625.1 GFXV01005779 MBW17584.1 GBYX01444793 JAO36627.1 JOJR01000128 RCN44473.1 CH933807 KRG03780.1 UYSL01006358 VDL67461.1 CAVP010058607 CDL94934.1 UZAF01017487 VDO42155.1 GBYX01444794 JAO36626.1 KN732949 KIH58590.1 KN716759 KJH41796.1 FN653053 CBY10183.1 ACPB03006481 EDW13484.2 GBYX01444792 JAO36628.1 KQ416708 KOF95609.1 GCES01164729 JAQ21593.1 KI660235 ETN76299.1 UYSU01036713 VDL98055.1 JARK01000389 EYC37453.1 EYC37454.1 DQ146416 AAZ75680.1 KPI94548.1 UZAO01078437 VDQ09963.1 JXLN01012619 KPM08691.1 UYRR01033424 VDK58736.1 JI169072 ADY43632.1 AYCK01010082 AYCK01010083 AYCK01010084 LSMT01000507 PFX16944.1 GDAI01000225 JAI17378.1 MH172491 QAA95898.1 FR916687 CDQ94049.1 AEMK02000017 KF669396 AIN44094.1 CM012445 RVE68758.1 RCN44479.1 HAEF01000612 SBR37994.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000245037
UP000235965
+ More
UP000027135 UP000038042 UP000268014 UP000008743 UP000183832 UP000192223 UP000007819 UP000252519 UP000009192 UP000271162 UP000025227 UP000038043 UP000053766 UP000015103 UP000007303 UP000002852 UP000053454 UP000261640 UP000261480 UP000050788 UP000275846 UP000265000 UP000024635 UP000050795 UP000050792 UP000261500 UP000036680 UP000028760 UP000264820 UP000225706 UP000265200 UP000265180 UP000001038 UP000193380 UP000008227 UP000035681
UP000027135 UP000038042 UP000268014 UP000008743 UP000183832 UP000192223 UP000007819 UP000252519 UP000009192 UP000271162 UP000025227 UP000038043 UP000053766 UP000015103 UP000007303 UP000002852 UP000053454 UP000261640 UP000261480 UP000050788 UP000275846 UP000265000 UP000024635 UP000050795 UP000050792 UP000261500 UP000036680 UP000028760 UP000264820 UP000225706 UP000265200 UP000265180 UP000001038 UP000193380 UP000008227 UP000035681
Pfam
Interpro
IPR017871
ABC_transporter_CS
+ More
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR026082 ABCA
IPR003959 ATPase_AAA_core
IPR031341 Methyltr_RsmF_N
IPR011023 Nop2p
IPR001678 MeTrfase_RsmB/NOP2
IPR029063 SAM-dependent_MTases
IPR018314 Fmu/NOL1/Nop2p_CS
IPR023273 RCMT_NOP2
IPR023267 RCMT
IPR020846 MFS_dom
IPR025302 DUF4162
IPR017994 P_trefoil_chordata
IPR000519 P_trefoil_dom
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR026082 ABCA
IPR003959 ATPase_AAA_core
IPR031341 Methyltr_RsmF_N
IPR011023 Nop2p
IPR001678 MeTrfase_RsmB/NOP2
IPR029063 SAM-dependent_MTases
IPR018314 Fmu/NOL1/Nop2p_CS
IPR023273 RCMT_NOP2
IPR023267 RCMT
IPR020846 MFS_dom
IPR025302 DUF4162
IPR017994 P_trefoil_chordata
IPR000519 P_trefoil_dom
ProteinModelPortal
A0A2W1BCS5
A0A194PTT1
A0A194R3K7
A0A2A4K545
A0A0F6PQM2
A0A212F3T3
+ More
A0A2P8YJ86 A0A1B6EFI2 A0A1B6DK03 A0A1B6JS44 A0A1B6FQX5 A0A1B6FLT5 A0A1B6F8C5 A0A1B6MC35 A0A1B6G0W6 A0A1B6IZN0 A0A1B6MPR0 A0A2J7RIF1 A0A1B6CKN8 A0A2J7RIF3 A0A067QMM5 A0A1B6GZC0 A0A1B6HI75 A0A0N4XAZ4 A0A0D2WPE2 E4Y7D9 A0A1J1IJX5 A0A1W4WY30 A0A2G9V2A8 A0A0A7ARI5 E4XQ17 A0A2S2P9P8 A0A1B6GC71 A0A1Y3AWT5 A0A2S2QUV9 A0A1B6HGH5 J9K9B5 A0A1B6I525 A0A2R7W837 A0A0S7H6F6 A0A2H8TTL0 A0A0S7HAW5 A0A368GJA2 A0A0Q9XF97 A0A3P7C2C3 W6NGB2 A0A158QNT6 A0A0S7HA71 A0A0N4XMX0 A0A0C2GBZ7 A0A0D8XHI3 E4XHW7 T1IBQ4 H3C1X8 B4KIX1 H3D169 A0A0S7H654 A0A3B5PPH9 A0A3B5PRM2 A0A0L8I2K3 A0A146MPG2 M3ZRP8 A0A3Q3RQP1 A0A3B3YAD3 A0A3Q3LC94 W2T430 A0A3B3WRC0 A0A183T5C2 A0A3Q2Q6M2 A0A016WEL3 A0A016WC99 Q3YB24 A0A194PNI5 A0A183WPN0 A0A132ACH8 A0A183QDX2 A0A3P6R8D8 A0A3B3V8X0 A0A0M3K8V1 F1L0H8 A0A087XBS7 A0A3Q2YND9 A0A2B4RF03 A0A3P9HY39 A0A0K8TT98 A0A3P9L2H4 A0A3B3H814 A0A3B3I8Q3 A0A3P9L2N4 A0A3B3WR13 A0A3P9HY72 A0A3R5SPL4 A0A060YX36 A0A287AQ53 A0A161ALT0 A0A3S2MX29 A0A368GMG1 A0A1A8L276 A0A0K0E8W1
A0A2P8YJ86 A0A1B6EFI2 A0A1B6DK03 A0A1B6JS44 A0A1B6FQX5 A0A1B6FLT5 A0A1B6F8C5 A0A1B6MC35 A0A1B6G0W6 A0A1B6IZN0 A0A1B6MPR0 A0A2J7RIF1 A0A1B6CKN8 A0A2J7RIF3 A0A067QMM5 A0A1B6GZC0 A0A1B6HI75 A0A0N4XAZ4 A0A0D2WPE2 E4Y7D9 A0A1J1IJX5 A0A1W4WY30 A0A2G9V2A8 A0A0A7ARI5 E4XQ17 A0A2S2P9P8 A0A1B6GC71 A0A1Y3AWT5 A0A2S2QUV9 A0A1B6HGH5 J9K9B5 A0A1B6I525 A0A2R7W837 A0A0S7H6F6 A0A2H8TTL0 A0A0S7HAW5 A0A368GJA2 A0A0Q9XF97 A0A3P7C2C3 W6NGB2 A0A158QNT6 A0A0S7HA71 A0A0N4XMX0 A0A0C2GBZ7 A0A0D8XHI3 E4XHW7 T1IBQ4 H3C1X8 B4KIX1 H3D169 A0A0S7H654 A0A3B5PPH9 A0A3B5PRM2 A0A0L8I2K3 A0A146MPG2 M3ZRP8 A0A3Q3RQP1 A0A3B3YAD3 A0A3Q3LC94 W2T430 A0A3B3WRC0 A0A183T5C2 A0A3Q2Q6M2 A0A016WEL3 A0A016WC99 Q3YB24 A0A194PNI5 A0A183WPN0 A0A132ACH8 A0A183QDX2 A0A3P6R8D8 A0A3B3V8X0 A0A0M3K8V1 F1L0H8 A0A087XBS7 A0A3Q2YND9 A0A2B4RF03 A0A3P9HY39 A0A0K8TT98 A0A3P9L2H4 A0A3B3H814 A0A3B3I8Q3 A0A3P9L2N4 A0A3B3WR13 A0A3P9HY72 A0A3R5SPL4 A0A060YX36 A0A287AQ53 A0A161ALT0 A0A3S2MX29 A0A368GMG1 A0A1A8L276 A0A0K0E8W1
PDB
5XJY
E-value=4.06554e-27,
Score=298
Ontologies
GO
PANTHER
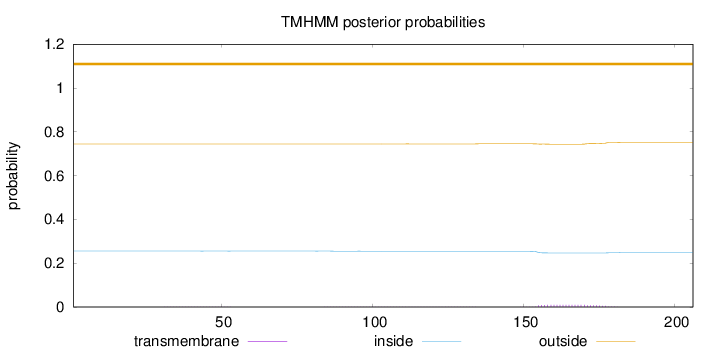
Topology
Length:
206
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23852
Exp number, first 60 AAs:
0.00529
Total prob of N-in:
0.25561
outside
1 - 206
Population Genetic Test Statistics
Pi
17.199091
Theta
21.028807
Tajima's D
-0.528113
CLR
11.238902
CSRT
0.237238138093095
Interpretation
Uncertain
