Gene
KWMTBOMO11725 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004187
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_A_member_3-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.683
Sequence
CDS
ATGCCAGTTCTCGTTTGGGTAAACCTGACCGCGTCGCACTTCCTGAACGTGCTGGCGGGCGGCGTGCTGTACGCGGGCGTGGTGTGCGGCGTGCTCACGGCGGACGTGGCGCTGTTCGAGACGCGCGCGGCCGCCGCCCTGCTGTGCGTCGGCCTGCTGCTGCACTTCGTCAGCGTCGTCGGGGCCGCCTTCGTCTGCAGCTACATCGTGAGAAGCACACAATACGTGGCGTCGGTGTCGGTGATCTCGTACGGGGTGCTGGCATACCCGATACACGCGCTGCGGGGGTTGACCATGCCCGGCGCGCTGAGGCTGCTGACGGGGCTGCTGCCGCACGTGCCGCTCGCCTGGCTGCTGGACGAGCTGAGCGTCTTGGACGGAAACGGTGAGTCGATCACAGTGTCGTCGATAATGAAGTCCCACACGGCCGTCAGCTACTCCGTGTTCGTGAGCCATCTGATGATGCTGGCCCAGATCCTAATATTCTTCTTCCTGAACTGGTATCTGTCTCTGGTCAGACCCGGAAAGTACGGACAAGCTTTACCCTGGTACTTCTTGTTTAAGAAACAATACTGGTCGAAGAACGAAATAGGGCCCAAGTTCGACTCGGACGCCGAGGAGCAGGTGCCGATAGTCAACGACAAGAAGTACTTCGAGGAGCCGCCGCAGAACGCCGAGGTTGGCATCCGGATCTGCAACGTGTCCAAGGTGTTCCAGAATCAGAAAGCCCTCGACAACGTGTCGCTGGATGTGTACAAGGGCGAGATCACCGTGCTGCTGGGACACAACGGCGCCGGCAAGACCACCCTCATGTCCATCATAACCGGTATGATGAGCGCGACCGAGGGTAAGGTGTACGTGAACGGCACGGAGACTGCCTCCCAGCAGAGGGAAGTCCGGAAGCACCTCGGGCTCTGCCCCCAGCACAACCTGTTCTTCCCGGACCTCACCGTGCTCGAACACGTCATGTTCTTCACTATGCTCAGGGGAACATCCTTCAGAGAAGCGAAAGCGTCTTCGAAAGCTCTTCTCGAACAGTTGGGTATCGCGGATAAAGCGAACAGCAACAGCTCCGAGCTGTCCGGCGGCATGATGAGGAGGCTGCAGCTGGCGTGCGCGCTCGCCGGCGACGCGGACGTCCTCATCCTGGACGAGCCCACGTCGGGGCTGGACGTGGAGACCAGGAGGAGCCTGTGGGACCTGTTGCTTAGCGTGCGAGGGTCGCGCACCGTGCTGCTGTCCACGCACTTCATGGAGGAGGCGGACGCGCTGGGTCACCGCGCCGCCGCGCTGCACCACGGCCGGCTGCGCTGCCACGCCTCCACCATGTACCTCAAGAAGGCCATCGGTACGGGATATCGGCTGTCGTTCACGACGATCGGGCTGCCCAACGAGGCGGCCATCACCGCGGCCGTCGCGTCGCTGGTCCCCGAAGTGACCCTCAAGGAGAAGTCGCTGAACACGCTCGCGTACAGTCTGCCGGCCGCGCACGCCCACAGGTTCCCGGACCTCTTCCTGGCGTTAGAGTCGAACCGATCTAAATTGGGCATCGACTCCATCGGCGTGGGCATCTCTACGCTGGAGGAAGTATTCCTGAAACTTTGCAGCGATGTCGACACGATCTTCGCCGACGCCAATGGTGACACCAGCGAAACTGCGGACCACTACCAGGAGCCGCAGAGTAAGAGGCTGACCGGCTTCCCCCTGTACCTCAGACAATTCAATGTCCTAATGAAAAGGCAGTTCAACTACATAATGTCCAAGAAACTCTCATTTTTCTTATTGCAAGTCCTGATACCTATGCTAGTGATCGGCCTGGTCACGGAGTCCGTGAACCACGACTACGGGTCCGCGCGGGCCGACGTCAGCGCGCCCATGACCCTCGACCTGTACAAGGACATCGACGGCCTCAAAGCTTTGTACAACGTGAACCAAGAGGGAGTCAACTTCAAAGCGATCACTAACGCCTACCCGCACGTCAAGTTTGAGGAAAATACAGACATCGTCAATACAACACTGGGCTACGCCAAGGGTGATCCAATGAAGTACATTAAATACGTTCTGGGTGTTGAACTTAATTCCACACATGCAAAGGCATTGTACACGACGACAATCCGGCACGCCGCTCCGATATCGGTGAACGTCCTGACGAACCTGCTGGCGACGCTGCACGCGCCGGCGGCGGGCGCGCGCACGCTCACCGCCGTCAACGAGCCCGTGCGCGCGGAGCTGGCGGCCGCACCCTCCTCCGCGCACTCCTCCGCGCCCTCCGCCAAGGACACTAAACAAGTCTTCGTCGTCCTCCTTTGGATATTTTTAGTGGTTTTCACGATTCTATCGACGACCATAAACGCGGTGTCGCTGCCGTGTGCGGAGCGGGAGTCGGGCGCGCGGCACACGCACGTCATGTCGGGCTGCCCGCCGGAGCTGCACTGGGCCTCCACGCTGCTGTTCCACCTGGCCGTCTGCACCGCGGCGCTGGTCGTCCCCACGCTGGTCATCGCGGCCACCGTCGAACGAGACCAGACCATCAACCAGCCTGGCTTCTTGCTAACGTTCTTCCTAATCTTGGAGCTCGGCGCTATGGCTTTCTTCGCGTTAATATATTTAGTGAGCTTCCACTTCAACGAGACGTCGGCCACTATCGTGCTATCCGCCATCGTTATTATATTCGGCATCTTCACGCCCTCAGTGAGCGTCATCTTCACGATCCTCGATGATGCGAAGGGTGTTACCTACTACGTGTTCTCGGTGATCAACTACGTGGTCCCGCCTCACACCGTGATGGTGGCGAGCGTGCTGGCGGGCATGCGGGCCGTGAACAACGCCATGTGCAAACTGGACTCGGAGTTCTGTTCCATCGCCATCGGGAGACAGATGACCACCGTTTCAAAGGATCTGGATCCGACGTGCTACCTATGCTTCAACGACATCGCCCCCACACGCTTCGTTATAATGCTGTTCGTTCAGTTCGCCTTTTTAATGAGCCTGGTGCTCCTTTCGGAGCGGGGCTGGCTGAACGGCTGCTTCGACGCGCTCATGAACCGCGCCTACCCCGCCGCGCCCCCCGCCGCCTCCCCCTCCGACCTCGTGCGCGCCGAGGCCGCTTACGTCAGCAAGGCGATCACACTGCGTACGTTTTCTTTTTTTTTTATTGCTAGAAGGGTGGACGAGCTCACAGCCCACCTGGTGTTAAGTGGTTACTGGAGCCCATAG
Protein
MPVLVWVNLTASHFLNVLAGGVLYAGVVCGVLTADVALFETRAAAALLCVGLLLHFVSVVGAAFVCSYIVRSTQYVASVSVISYGVLAYPIHALRGLTMPGALRLLTGLLPHVPLAWLLDELSVLDGNGESITVSSIMKSHTAVSYSVFVSHLMMLAQILIFFFLNWYLSLVRPGKYGQALPWYFLFKKQYWSKNEIGPKFDSDAEEQVPIVNDKKYFEEPPQNAEVGIRICNVSKVFQNQKALDNVSLDVYKGEITVLLGHNGAGKTTLMSIITGMMSATEGKVYVNGTETASQQREVRKHLGLCPQHNLFFPDLTVLEHVMFFTMLRGTSFREAKASSKALLEQLGIADKANSNSSELSGGMMRRLQLACALAGDADVLILDEPTSGLDVETRRSLWDLLLSVRGSRTVLLSTHFMEEADALGHRAAALHHGRLRCHASTMYLKKAIGTGYRLSFTTIGLPNEAAITAAVASLVPEVTLKEKSLNTLAYSLPAAHAHRFPDLFLALESNRSKLGIDSIGVGISTLEEVFLKLCSDVDTIFADANGDTSETADHYQEPQSKRLTGFPLYLRQFNVLMKRQFNYIMSKKLSFFLLQVLIPMLVIGLVTESVNHDYGSARADVSAPMTLDLYKDIDGLKALYNVNQEGVNFKAITNAYPHVKFEENTDIVNTTLGYAKGDPMKYIKYVLGVELNSTHAKALYTTTIRHAAPISVNVLTNLLATLHAPAAGARTLTAVNEPVRAELAAAPSSAHSSAPSAKDTKQVFVVLLWIFLVVFTILSTTINAVSLPCAERESGARHTHVMSGCPPELHWASTLLFHLAVCTAALVVPTLVIAATVERDQTINQPGFLLTFFLILELGAMAFFALIYLVSFHFNETSATIVLSAIVIIFGIFTPSVSVIFTILDDAKGVTYYVFSVINYVVPPHTVMVASVLAGMRAVNNAMCKLDSEFCSIAIGRQMTTVSKDLDPTCYLCFNDIAPTRFVIMLFVQFAFLMSLVLLSERGWLNGCFDALMNRAYPAAPPAASPSDLVRAEAAYVSKAITLRTFSFFFIARRVDELTAHLVLSGYWSP
Summary
Uniprot
A0A2W1BCS5
A0A2A4K545
A0A194PTT1
A0A212F3T3
A0A194R3K7
A0A2J7RIF3
+ More
A0A1B6GJX2 A0A1B6MNY8 A0A067QMM5 A0A1B6GC71 A0A1B6GKN0 A0A1B6HXX1 A0A1B6FUV8 A0A1B6L5W6 A0A2A4JHG4 A0A0A7AS05 T1G4X2 A0A0A9VSY3 A0A161ALT0 A0A1B6GZC0 A0A1W4WY30 A0A1B6GMP0 A0A0A9VVV1 A0A146KLZ7 A0A0A9W0E8 A0A1B6D8R9 T1JGM6 A0A0L8GMY3 A0A210QSG5 A0A3S3QLI1 B4L8N3 A0A1D2NL00 A0A1I8Q220 A0A1Y1JV91 Q16LB6 A0A0L0BW92 F1M2Q5 E0VMF5 M0R7Y5 A0A226DL54 W8AEW5 A0A131Z7N0 A0A0A1X7V3 A0A1Q3FHX7 A0A1Q3FHW2 A0A0A1WS91 A0A1Q3FHU1 A0A1S3A252 A0A2H4KC39 A0A1Q3FHY8 A0A0K8U924 A0A182GWA1 L8J5Q4 K9IQF6 G1PVG9 F6Z1P2 A0A2H4TEN0 A0A182GWA4 Q16LB7 L5LVJ4 A0A1S4FWY8 I3MKK6 A0A1B0DJU5 A0A131Z7J7 E9PWH4 Q6XBG2 E1BCR1
A0A1B6GJX2 A0A1B6MNY8 A0A067QMM5 A0A1B6GC71 A0A1B6GKN0 A0A1B6HXX1 A0A1B6FUV8 A0A1B6L5W6 A0A2A4JHG4 A0A0A7AS05 T1G4X2 A0A0A9VSY3 A0A161ALT0 A0A1B6GZC0 A0A1W4WY30 A0A1B6GMP0 A0A0A9VVV1 A0A146KLZ7 A0A0A9W0E8 A0A1B6D8R9 T1JGM6 A0A0L8GMY3 A0A210QSG5 A0A3S3QLI1 B4L8N3 A0A1D2NL00 A0A1I8Q220 A0A1Y1JV91 Q16LB6 A0A0L0BW92 F1M2Q5 E0VMF5 M0R7Y5 A0A226DL54 W8AEW5 A0A131Z7N0 A0A0A1X7V3 A0A1Q3FHX7 A0A1Q3FHW2 A0A0A1WS91 A0A1Q3FHU1 A0A1S3A252 A0A2H4KC39 A0A1Q3FHY8 A0A0K8U924 A0A182GWA1 L8J5Q4 K9IQF6 G1PVG9 F6Z1P2 A0A2H4TEN0 A0A182GWA4 Q16LB7 L5LVJ4 A0A1S4FWY8 I3MKK6 A0A1B0DJU5 A0A131Z7J7 E9PWH4 Q6XBG2 E1BCR1
Pubmed
EMBL
KZ150130
PZC73059.1
NWSH01000149
PCG79038.1
KQ459598
KPI94545.1
+ More
AGBW02010485 OWR48406.1 KQ460949 KPJ10426.1 NEVH01003500 PNF40610.1 GECZ01007017 JAS62752.1 GEBQ01002309 JAT37668.1 KK853244 KDR09441.1 GECZ01009744 JAS60025.1 GECZ01006770 JAS62999.1 GECU01028202 JAS79504.1 GECZ01015840 JAS53929.1 GEBQ01020917 JAT19060.1 NWSH01001571 PCG70832.1 KF906266 AHK05631.1 AMQM01005314 KB096900 ESO00555.1 GBHO01045258 JAF98345.1 KF669396 AIN44094.1 GECZ01001999 JAS67770.1 GECZ01006109 JAS63660.1 GBHO01045261 GBRD01003190 JAF98342.1 JAG62631.1 GDHC01021046 JAP97582.1 GBHO01045259 JAF98344.1 GEDC01015204 JAS22094.1 JH432211 KQ421117 KOF78343.1 NEDP02002179 OWF51665.1 NCKU01002032 NCKU01000808 RWS10623.1 RWS14067.1 CH933815 EDW08008.2 LJIJ01000013 ODN05940.1 GEZM01100130 JAV53003.1 CH477911 EAT35113.1 JRES01001244 KNC24298.1 AABR07005599 AABR07005600 AABR07005601 AABR07005602 AABR07005603 AABR07005604 DS235307 EEB14561.1 LNIX01000016 OXA45953.1 GAMC01019799 GAMC01019797 GAMC01019795 JAB86760.1 GEDV01001233 JAP87324.1 GBXI01007301 GBXI01005702 JAD06991.1 JAD08590.1 GFDL01007897 JAV27148.1 GFDL01007895 JAV27150.1 GBXI01012776 GBXI01008150 JAD01516.1 JAD06142.1 GFDL01007909 JAV27136.1 KY484773 ASS36015.1 GFDL01007896 JAV27149.1 GDHF01029484 GDHF01021855 GDHF01018869 GDHF01001688 JAI22830.1 JAI30459.1 JAI33445.1 JAI50626.1 JXUM01020746 KQ560580 KXJ81818.1 JH880268 ELR62954.1 GABZ01003538 JAA49987.1 AAPE02041718 KY849620 ATY74505.1 JXUM01020756 JXUM01020757 KXJ81821.1 EAT35112.1 KB107323 ELK30082.1 AGTP01095349 AJVK01001087 GEDV01001234 JAP87323.1 AC126434 AC134434 BC141350 BC141351 AY243471 AAI41351.1 AAP73045.1 GJ062847 DAA15550.1
AGBW02010485 OWR48406.1 KQ460949 KPJ10426.1 NEVH01003500 PNF40610.1 GECZ01007017 JAS62752.1 GEBQ01002309 JAT37668.1 KK853244 KDR09441.1 GECZ01009744 JAS60025.1 GECZ01006770 JAS62999.1 GECU01028202 JAS79504.1 GECZ01015840 JAS53929.1 GEBQ01020917 JAT19060.1 NWSH01001571 PCG70832.1 KF906266 AHK05631.1 AMQM01005314 KB096900 ESO00555.1 GBHO01045258 JAF98345.1 KF669396 AIN44094.1 GECZ01001999 JAS67770.1 GECZ01006109 JAS63660.1 GBHO01045261 GBRD01003190 JAF98342.1 JAG62631.1 GDHC01021046 JAP97582.1 GBHO01045259 JAF98344.1 GEDC01015204 JAS22094.1 JH432211 KQ421117 KOF78343.1 NEDP02002179 OWF51665.1 NCKU01002032 NCKU01000808 RWS10623.1 RWS14067.1 CH933815 EDW08008.2 LJIJ01000013 ODN05940.1 GEZM01100130 JAV53003.1 CH477911 EAT35113.1 JRES01001244 KNC24298.1 AABR07005599 AABR07005600 AABR07005601 AABR07005602 AABR07005603 AABR07005604 DS235307 EEB14561.1 LNIX01000016 OXA45953.1 GAMC01019799 GAMC01019797 GAMC01019795 JAB86760.1 GEDV01001233 JAP87324.1 GBXI01007301 GBXI01005702 JAD06991.1 JAD08590.1 GFDL01007897 JAV27148.1 GFDL01007895 JAV27150.1 GBXI01012776 GBXI01008150 JAD01516.1 JAD06142.1 GFDL01007909 JAV27136.1 KY484773 ASS36015.1 GFDL01007896 JAV27149.1 GDHF01029484 GDHF01021855 GDHF01018869 GDHF01001688 JAI22830.1 JAI30459.1 JAI33445.1 JAI50626.1 JXUM01020746 KQ560580 KXJ81818.1 JH880268 ELR62954.1 GABZ01003538 JAA49987.1 AAPE02041718 KY849620 ATY74505.1 JXUM01020756 JXUM01020757 KXJ81821.1 EAT35112.1 KB107323 ELK30082.1 AGTP01095349 AJVK01001087 GEDV01001234 JAP87323.1 AC126434 AC134434 BC141350 BC141351 AY243471 AAI41351.1 AAP73045.1 GJ062847 DAA15550.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000235965
UP000027135
+ More
UP000015101 UP000192223 UP000053454 UP000242188 UP000285301 UP000009192 UP000094527 UP000095300 UP000008820 UP000037069 UP000002494 UP000009046 UP000198287 UP000079721 UP000069940 UP000249989 UP000001074 UP000002281 UP000005215 UP000092462 UP000000589 UP000009136
UP000015101 UP000192223 UP000053454 UP000242188 UP000285301 UP000009192 UP000094527 UP000095300 UP000008820 UP000037069 UP000002494 UP000009046 UP000198287 UP000079721 UP000069940 UP000249989 UP000001074 UP000002281 UP000005215 UP000092462 UP000000589 UP000009136
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2W1BCS5
A0A2A4K545
A0A194PTT1
A0A212F3T3
A0A194R3K7
A0A2J7RIF3
+ More
A0A1B6GJX2 A0A1B6MNY8 A0A067QMM5 A0A1B6GC71 A0A1B6GKN0 A0A1B6HXX1 A0A1B6FUV8 A0A1B6L5W6 A0A2A4JHG4 A0A0A7AS05 T1G4X2 A0A0A9VSY3 A0A161ALT0 A0A1B6GZC0 A0A1W4WY30 A0A1B6GMP0 A0A0A9VVV1 A0A146KLZ7 A0A0A9W0E8 A0A1B6D8R9 T1JGM6 A0A0L8GMY3 A0A210QSG5 A0A3S3QLI1 B4L8N3 A0A1D2NL00 A0A1I8Q220 A0A1Y1JV91 Q16LB6 A0A0L0BW92 F1M2Q5 E0VMF5 M0R7Y5 A0A226DL54 W8AEW5 A0A131Z7N0 A0A0A1X7V3 A0A1Q3FHX7 A0A1Q3FHW2 A0A0A1WS91 A0A1Q3FHU1 A0A1S3A252 A0A2H4KC39 A0A1Q3FHY8 A0A0K8U924 A0A182GWA1 L8J5Q4 K9IQF6 G1PVG9 F6Z1P2 A0A2H4TEN0 A0A182GWA4 Q16LB7 L5LVJ4 A0A1S4FWY8 I3MKK6 A0A1B0DJU5 A0A131Z7J7 E9PWH4 Q6XBG2 E1BCR1
A0A1B6GJX2 A0A1B6MNY8 A0A067QMM5 A0A1B6GC71 A0A1B6GKN0 A0A1B6HXX1 A0A1B6FUV8 A0A1B6L5W6 A0A2A4JHG4 A0A0A7AS05 T1G4X2 A0A0A9VSY3 A0A161ALT0 A0A1B6GZC0 A0A1W4WY30 A0A1B6GMP0 A0A0A9VVV1 A0A146KLZ7 A0A0A9W0E8 A0A1B6D8R9 T1JGM6 A0A0L8GMY3 A0A210QSG5 A0A3S3QLI1 B4L8N3 A0A1D2NL00 A0A1I8Q220 A0A1Y1JV91 Q16LB6 A0A0L0BW92 F1M2Q5 E0VMF5 M0R7Y5 A0A226DL54 W8AEW5 A0A131Z7N0 A0A0A1X7V3 A0A1Q3FHX7 A0A1Q3FHW2 A0A0A1WS91 A0A1Q3FHU1 A0A1S3A252 A0A2H4KC39 A0A1Q3FHY8 A0A0K8U924 A0A182GWA1 L8J5Q4 K9IQF6 G1PVG9 F6Z1P2 A0A2H4TEN0 A0A182GWA4 Q16LB7 L5LVJ4 A0A1S4FWY8 I3MKK6 A0A1B0DJU5 A0A131Z7J7 E9PWH4 Q6XBG2 E1BCR1
PDB
5XJY
E-value=3.84359e-72,
Score=694
Ontologies
GO
PANTHER
Topology
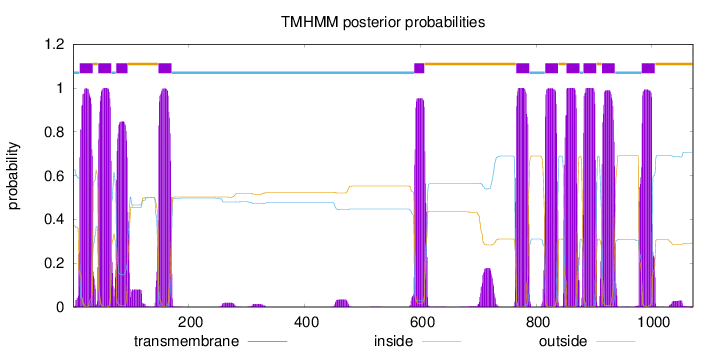
Length:
1071
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
247.726720000001
Exp number, first 60 AAs:
39.14472
Total prob of N-in:
0.62841
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 43
TMhelix
44 - 66
inside
67 - 74
TMhelix
75 - 94
outside
95 - 147
TMhelix
148 - 170
inside
171 - 589
TMhelix
590 - 607
outside
608 - 765
TMhelix
766 - 788
inside
789 - 815
TMhelix
816 - 838
outside
839 - 852
TMhelix
853 - 875
inside
876 - 881
TMhelix
882 - 904
outside
905 - 913
TMhelix
914 - 936
inside
937 - 982
TMhelix
983 - 1005
outside
1006 - 1071
Population Genetic Test Statistics
Pi
22.687601
Theta
24.159217
Tajima's D
-1.385914
CLR
1.202744
CSRT
0.0747962601869906
Interpretation
Uncertain
