Gene
KWMTBOMO11720
Pre Gene Modal
BGIBMGA004183
Annotation
PREDICTED:_uncharacterized_protein_LOC101736310_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.048
Sequence
CDS
ATGGAAGTATACTCTGAGTTCTGTGGGTTCTGTCTGGCCACGGGCAACGCTATTAGGAAGGGGAAATTGGAAATCGGACAAGATCGAATCGCGATGGTCAAAGCCTTAGCCGATAACATCACTGCTTTATCAAGATTTAAAGATAAAAGTGAACCCGTATTTCTATTCATATTTAAAGGTAAATTAACAAGAGCAATGTTTGGCGCCAACGGAATAGAGCTGTGCCGTATCATAGAGGAAGAGATGGGGCTCATGCAGCGGGAAATAGACACGGGAATCGTGAGGAAGAAGCAAGAGATCACGGAATTATTACCCGAGGAATCTGTTTGTCAATAG
Protein
MEVYSEFCGFCLATGNAIRKGKLEIGQDRIAMVKALADNITALSRFKDKSEPVFLFIFKGKLTRAMFGANGIELCRIIEEEMGLMQREIDTGIVRKKQEITELLPEESVCQ
Summary
Similarity
Belongs to the NDK family.
Uniprot
A0A194QY11
A0A194PTT6
A0A212F1G5
A0A2H1V6I7
A0A2A4K459
A0A0L7LB95
+ More
A0A336LND6 U4TZR8 A0A139WH38 W8B8C6 A0A0A1XPB8 A0A0K8W067 A0A034VR51 A0A0K8UQ08 T1GUM7 A0A1Y1KSD8 A0A1B6CN68 A0A067R9F7 A0A1B6H9V5 A0A084WHX9 A0A1B6CPK3 A0A1B6CSL6 A0A194R6K0 W5JN69 A0A182J572 A0A182R0E8 A0A182MYI7 A0A182FU39 A0A182MWZ6 A0A182Y1U2 A0A182W974 A0A3Q0II17 A0A182R5F4 Q172W6 Q7Q063 A0A182XD78 A0A182UR36 A0A1S4H9Y2 A0A182JNZ3 A0A182UKB7 A0A182KSP6 A0A182HQ17 A0A182PBQ3 A0A182GMV1 A0A1I8MHT2 A0A0L7LA20 A0A1Q3FT98 A0A1B6LQG4 A0A0L0CRZ1 A0A1I8N7U1 B0WTM5 A0A194PYQ6 A0A146KSJ8 A0A1B6F5V9 A0A1I8PLU9 A0A212F7X0 A0A2S2NCL2 B4JIQ9 A0A310S7E1 A0A2H1WP09 A0A0A9XKI7 A0A182SYK8 B4JK05 A0A0L7RIX8 A0A2A4K7L4 A0A2W1BU80 B3N0Q9 B4H0Y9 A0A1I8P0W3 A0A0A9WKZ1 A0A0C9RLJ0 N6U3E6 A0A1A9UG81 A0A3B4E009 K7JF81 A0A3B4CF31 X1WW09 A0A3B0K3E2 A0A3S2LJE5 A0A1A8BLT7 B4L4Y5 A0A2S2RB45 B3NXN0 A0A1B0FPJ9 B4L6D8 A0A1W4W660 B5DNI0 B4M233 B4NDU9 E2B7A0 B4PZF5 A0A310SLQ0 A0A3B0K524 B3MQB0 A0A158NIJ0 A0A151HZC7 A0A1A9WQ32 A0A3Q4HUF3
A0A336LND6 U4TZR8 A0A139WH38 W8B8C6 A0A0A1XPB8 A0A0K8W067 A0A034VR51 A0A0K8UQ08 T1GUM7 A0A1Y1KSD8 A0A1B6CN68 A0A067R9F7 A0A1B6H9V5 A0A084WHX9 A0A1B6CPK3 A0A1B6CSL6 A0A194R6K0 W5JN69 A0A182J572 A0A182R0E8 A0A182MYI7 A0A182FU39 A0A182MWZ6 A0A182Y1U2 A0A182W974 A0A3Q0II17 A0A182R5F4 Q172W6 Q7Q063 A0A182XD78 A0A182UR36 A0A1S4H9Y2 A0A182JNZ3 A0A182UKB7 A0A182KSP6 A0A182HQ17 A0A182PBQ3 A0A182GMV1 A0A1I8MHT2 A0A0L7LA20 A0A1Q3FT98 A0A1B6LQG4 A0A0L0CRZ1 A0A1I8N7U1 B0WTM5 A0A194PYQ6 A0A146KSJ8 A0A1B6F5V9 A0A1I8PLU9 A0A212F7X0 A0A2S2NCL2 B4JIQ9 A0A310S7E1 A0A2H1WP09 A0A0A9XKI7 A0A182SYK8 B4JK05 A0A0L7RIX8 A0A2A4K7L4 A0A2W1BU80 B3N0Q9 B4H0Y9 A0A1I8P0W3 A0A0A9WKZ1 A0A0C9RLJ0 N6U3E6 A0A1A9UG81 A0A3B4E009 K7JF81 A0A3B4CF31 X1WW09 A0A3B0K3E2 A0A3S2LJE5 A0A1A8BLT7 B4L4Y5 A0A2S2RB45 B3NXN0 A0A1B0FPJ9 B4L6D8 A0A1W4W660 B5DNI0 B4M233 B4NDU9 E2B7A0 B4PZF5 A0A310SLQ0 A0A3B0K524 B3MQB0 A0A158NIJ0 A0A151HZC7 A0A1A9WQ32 A0A3Q4HUF3
Pubmed
EMBL
KQ460949
KPJ10428.1
KQ459598
KPI94550.1
AGBW02010879
OWR47585.1
+ More
ODYU01000925 SOQ36427.1 NWSH01000149 PCG79037.1 JTDY01001878 KOB72654.1 UFQT01000028 SSX18183.1 KB630709 ERL83801.1 KQ971343 KYB27226.1 GAMC01020536 GAMC01020534 JAB86019.1 GBXI01001889 JAD12403.1 GDHF01007756 JAI44558.1 GAKP01013998 JAC44954.1 GDHF01023643 JAI28671.1 CAQQ02171786 GEZM01077275 JAV63341.1 GEDC01022445 JAS14853.1 KK852609 KDR20317.1 GECU01036259 JAS71447.1 ATLV01023896 KE525347 KFB49823.1 GEDC01021909 JAS15389.1 GEDC01021055 JAS16243.1 KQ460644 KPJ13292.1 ADMH02001044 ETN64335.1 AXCP01006761 AXCN02001041 AXCM01002401 CH477430 EAT41063.1 AAAB01008986 EAA00251.4 APCN01003288 JXUM01014260 KQ560398 KXJ82621.1 JTDY01002051 KOB72234.1 GFDL01004248 JAV30797.1 GEBQ01014032 JAT25945.1 JRES01000005 KNC34991.1 DS232089 EDS34478.1 KQ459584 KPI98461.1 GDHC01019146 JAP99482.1 GECZ01024204 JAS45565.1 AGBW02009824 OWR49832.1 GGMR01002301 MBY14920.1 CH916370 EDW00506.1 KQ766110 OAD53747.1 ODYU01009988 SOQ54768.1 GBHO01023115 JAG20489.1 EDV99907.1 KQ414582 KOC70902.1 NWSH01000047 PCG80221.1 KZ149934 PZC77215.1 CH902644 EDV34788.1 CH479201 EDW29905.1 GBHO01035523 JAG08081.1 GBYB01007806 JAG77573.1 APGK01044156 KB741020 ENN75141.1 AAZX01000586 ABLF02032254 ABLF02032259 OUUW01000015 SPP88757.1 RSAL01000001 RVE55167.1 HADZ01003655 SBP67596.1 CH933810 EDW07613.1 GGMS01017971 MBY87174.1 CH954180 EDV47331.2 CCAG010022945 CH933812 EDW05934.2 CH379064 EDY72988.2 CH940651 EDW65737.1 CH964239 EDW81918.1 GL446149 EFN88461.1 CM000162 EDX02111.1 KQ763877 OAD54657.1 OUUW01000020 SPP89337.1 CH902621 EDV44536.1 ADTU01016761 KQ976705 KYM77076.1
ODYU01000925 SOQ36427.1 NWSH01000149 PCG79037.1 JTDY01001878 KOB72654.1 UFQT01000028 SSX18183.1 KB630709 ERL83801.1 KQ971343 KYB27226.1 GAMC01020536 GAMC01020534 JAB86019.1 GBXI01001889 JAD12403.1 GDHF01007756 JAI44558.1 GAKP01013998 JAC44954.1 GDHF01023643 JAI28671.1 CAQQ02171786 GEZM01077275 JAV63341.1 GEDC01022445 JAS14853.1 KK852609 KDR20317.1 GECU01036259 JAS71447.1 ATLV01023896 KE525347 KFB49823.1 GEDC01021909 JAS15389.1 GEDC01021055 JAS16243.1 KQ460644 KPJ13292.1 ADMH02001044 ETN64335.1 AXCP01006761 AXCN02001041 AXCM01002401 CH477430 EAT41063.1 AAAB01008986 EAA00251.4 APCN01003288 JXUM01014260 KQ560398 KXJ82621.1 JTDY01002051 KOB72234.1 GFDL01004248 JAV30797.1 GEBQ01014032 JAT25945.1 JRES01000005 KNC34991.1 DS232089 EDS34478.1 KQ459584 KPI98461.1 GDHC01019146 JAP99482.1 GECZ01024204 JAS45565.1 AGBW02009824 OWR49832.1 GGMR01002301 MBY14920.1 CH916370 EDW00506.1 KQ766110 OAD53747.1 ODYU01009988 SOQ54768.1 GBHO01023115 JAG20489.1 EDV99907.1 KQ414582 KOC70902.1 NWSH01000047 PCG80221.1 KZ149934 PZC77215.1 CH902644 EDV34788.1 CH479201 EDW29905.1 GBHO01035523 JAG08081.1 GBYB01007806 JAG77573.1 APGK01044156 KB741020 ENN75141.1 AAZX01000586 ABLF02032254 ABLF02032259 OUUW01000015 SPP88757.1 RSAL01000001 RVE55167.1 HADZ01003655 SBP67596.1 CH933810 EDW07613.1 GGMS01017971 MBY87174.1 CH954180 EDV47331.2 CCAG010022945 CH933812 EDW05934.2 CH379064 EDY72988.2 CH940651 EDW65737.1 CH964239 EDW81918.1 GL446149 EFN88461.1 CM000162 EDX02111.1 KQ763877 OAD54657.1 OUUW01000020 SPP89337.1 CH902621 EDV44536.1 ADTU01016761 KQ976705 KYM77076.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000037510
UP000030742
+ More
UP000007266 UP000015102 UP000027135 UP000030765 UP000000673 UP000075880 UP000075886 UP000075884 UP000069272 UP000075883 UP000076408 UP000075920 UP000079169 UP000075900 UP000008820 UP000007062 UP000076407 UP000075903 UP000075881 UP000075902 UP000075882 UP000075840 UP000075885 UP000069940 UP000249989 UP000095301 UP000037069 UP000002320 UP000095300 UP000001070 UP000075901 UP000053825 UP000007801 UP000008744 UP000019118 UP000078200 UP000261440 UP000002358 UP000007819 UP000268350 UP000283053 UP000009192 UP000008711 UP000092444 UP000192223 UP000001819 UP000008792 UP000007798 UP000008237 UP000002282 UP000005205 UP000078540 UP000091820 UP000261580
UP000007266 UP000015102 UP000027135 UP000030765 UP000000673 UP000075880 UP000075886 UP000075884 UP000069272 UP000075883 UP000076408 UP000075920 UP000079169 UP000075900 UP000008820 UP000007062 UP000076407 UP000075903 UP000075881 UP000075902 UP000075882 UP000075840 UP000075885 UP000069940 UP000249989 UP000095301 UP000037069 UP000002320 UP000095300 UP000001070 UP000075901 UP000053825 UP000007801 UP000008744 UP000019118 UP000078200 UP000261440 UP000002358 UP000007819 UP000268350 UP000283053 UP000009192 UP000008711 UP000092444 UP000192223 UP000001819 UP000008792 UP000007798 UP000008237 UP000002282 UP000005205 UP000078540 UP000091820 UP000261580
Interpro
Gene 3D
ProteinModelPortal
A0A194QY11
A0A194PTT6
A0A212F1G5
A0A2H1V6I7
A0A2A4K459
A0A0L7LB95
+ More
A0A336LND6 U4TZR8 A0A139WH38 W8B8C6 A0A0A1XPB8 A0A0K8W067 A0A034VR51 A0A0K8UQ08 T1GUM7 A0A1Y1KSD8 A0A1B6CN68 A0A067R9F7 A0A1B6H9V5 A0A084WHX9 A0A1B6CPK3 A0A1B6CSL6 A0A194R6K0 W5JN69 A0A182J572 A0A182R0E8 A0A182MYI7 A0A182FU39 A0A182MWZ6 A0A182Y1U2 A0A182W974 A0A3Q0II17 A0A182R5F4 Q172W6 Q7Q063 A0A182XD78 A0A182UR36 A0A1S4H9Y2 A0A182JNZ3 A0A182UKB7 A0A182KSP6 A0A182HQ17 A0A182PBQ3 A0A182GMV1 A0A1I8MHT2 A0A0L7LA20 A0A1Q3FT98 A0A1B6LQG4 A0A0L0CRZ1 A0A1I8N7U1 B0WTM5 A0A194PYQ6 A0A146KSJ8 A0A1B6F5V9 A0A1I8PLU9 A0A212F7X0 A0A2S2NCL2 B4JIQ9 A0A310S7E1 A0A2H1WP09 A0A0A9XKI7 A0A182SYK8 B4JK05 A0A0L7RIX8 A0A2A4K7L4 A0A2W1BU80 B3N0Q9 B4H0Y9 A0A1I8P0W3 A0A0A9WKZ1 A0A0C9RLJ0 N6U3E6 A0A1A9UG81 A0A3B4E009 K7JF81 A0A3B4CF31 X1WW09 A0A3B0K3E2 A0A3S2LJE5 A0A1A8BLT7 B4L4Y5 A0A2S2RB45 B3NXN0 A0A1B0FPJ9 B4L6D8 A0A1W4W660 B5DNI0 B4M233 B4NDU9 E2B7A0 B4PZF5 A0A310SLQ0 A0A3B0K524 B3MQB0 A0A158NIJ0 A0A151HZC7 A0A1A9WQ32 A0A3Q4HUF3
A0A336LND6 U4TZR8 A0A139WH38 W8B8C6 A0A0A1XPB8 A0A0K8W067 A0A034VR51 A0A0K8UQ08 T1GUM7 A0A1Y1KSD8 A0A1B6CN68 A0A067R9F7 A0A1B6H9V5 A0A084WHX9 A0A1B6CPK3 A0A1B6CSL6 A0A194R6K0 W5JN69 A0A182J572 A0A182R0E8 A0A182MYI7 A0A182FU39 A0A182MWZ6 A0A182Y1U2 A0A182W974 A0A3Q0II17 A0A182R5F4 Q172W6 Q7Q063 A0A182XD78 A0A182UR36 A0A1S4H9Y2 A0A182JNZ3 A0A182UKB7 A0A182KSP6 A0A182HQ17 A0A182PBQ3 A0A182GMV1 A0A1I8MHT2 A0A0L7LA20 A0A1Q3FT98 A0A1B6LQG4 A0A0L0CRZ1 A0A1I8N7U1 B0WTM5 A0A194PYQ6 A0A146KSJ8 A0A1B6F5V9 A0A1I8PLU9 A0A212F7X0 A0A2S2NCL2 B4JIQ9 A0A310S7E1 A0A2H1WP09 A0A0A9XKI7 A0A182SYK8 B4JK05 A0A0L7RIX8 A0A2A4K7L4 A0A2W1BU80 B3N0Q9 B4H0Y9 A0A1I8P0W3 A0A0A9WKZ1 A0A0C9RLJ0 N6U3E6 A0A1A9UG81 A0A3B4E009 K7JF81 A0A3B4CF31 X1WW09 A0A3B0K3E2 A0A3S2LJE5 A0A1A8BLT7 B4L4Y5 A0A2S2RB45 B3NXN0 A0A1B0FPJ9 B4L6D8 A0A1W4W660 B5DNI0 B4M233 B4NDU9 E2B7A0 B4PZF5 A0A310SLQ0 A0A3B0K524 B3MQB0 A0A158NIJ0 A0A151HZC7 A0A1A9WQ32 A0A3Q4HUF3
Ontologies
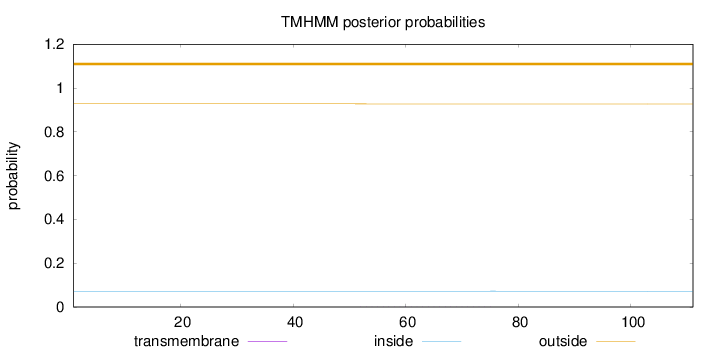
Topology
Length:
111
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01226
Exp number, first 60 AAs:
0.00477
Total prob of N-in:
0.07203
outside
1 - 111
Population Genetic Test Statistics
Pi
28.488112
Theta
27.464673
Tajima's D
0.244982
CLR
0.477281
CSRT
0.443377831108445
Interpretation
Uncertain
