Pre Gene Modal
BGIBMGA004178
Annotation
PREDICTED:_oxysterol-binding_protein-related_protein_9_[Bombyx_mori]
Full name
Oxysterol-binding protein
Location in the cell
Cytoplasmic Reliability : 1.814 Nuclear Reliability : 2.137
Sequence
CDS
ATGTCTTGTAACGTTAGCAAGAATCATCCGGCGGAGGGCACGCCCGAGCAAGACTCGGCGCAGCCCGCGACCGTCGACCTGCCTCGCACTAGAGATGGAGAGATCGACTATGATGCTCTCTACGAGGACGAATCGGACAGTGACCTCGGATCGATGGAGAACCACGGCTCGGTTGTCACGCATCTCCTGTCGCAGGTCAAGATCGGAATGGATCTCACAAAGGTTGTGCTCCCGACTTTTATACTGGAAAGACGATCGCTATTGGAAATGTACGCGGATTCGTTCGCTCATCCCGACCAATTTATAAAGATAGTAGAGTTGCCGACGCCGCGCGAAAGAATGGTGCAAGTGATCCGCTGGTACCTGAGCTCGTATCACGCCGGTCGCAAGTCGCAAGTCGCCAAGAAACCCTACAACCCCATACTGGGGGAGGTGTTCCGCTGTCACTGGGACCTGGACGGTGCACTCGATAACCCCAATGATAAGCAAGAGGTAGGGGACGGTCCGGTACCGTGGTGCAGTCCGGACCAGTTGTCGTTCGTGGCCGAGCAAGTGTCCCACCATCCTCCCATATCGGCGTTCTACGCGGAGCACGTCAACAAACGCATCCAGTTCGAAGCCTGGGTGTGGACGAAGAGCAAATTCCTCGGCCTGTCCATAGGAGTCCACAACATCGGCAAGGGCACGGTCACGCTCATCGATCTCGGAGAGGAGTACACTTTGACATTCCCCAATGGTTACGGCAGATCAATATTAACGGTACCGTGGATCGAACTGGGCGGCTCTGTGGTCATCGAGTGCGTGCAAACCGGCCACAAAGCCAACATAGAGTTCCTAACGAAGCCATTCTACGGCGGCAAGAAGCACAGGGTGACCTGCGAGATATTCGCCGGCGCTGAGAAGAAGCCGTACTACACCGCGCAAGGCGAGTGGAACACCAGGATGGATGGGAGGTGGACCGACTCAGGGCGGTCGGAGATCCTGTTCGAAGTGAGCAACATGAAGCCGCGGCGCAAGCGCGTGGCGCCCGTGAGCCGGCAGCTGCCGCACGAGTCGCGCCGCGTGTGGCGCCACGTGACGTGCGCCCTGCGCGCCGCCGACTGCGACGCCGCCACGCTCGCCAAGCGACACGTGGAGCAGGCGCAGCGGGACGCAGTCAAGCACCGCGAGGCCAGCGGGGAGAAGTGGCAGACGCAGCTTTTCAGTCCCAGGATCGAAGAAGGCTGGGAGTACAAGTCGCCTCTCAAGAACCGCAACGTGCCCTGCAAATCGCCCCAACACAAGCACTCTGAGAACGACAATACGAGATAA
Protein
MSCNVSKNHPAEGTPEQDSAQPATVDLPRTRDGEIDYDALYEDESDSDLGSMENHGSVVTHLLSQVKIGMDLTKVVLPTFILERRSLLEMYADSFAHPDQFIKIVELPTPRERMVQVIRWYLSSYHAGRKSQVAKKPYNPILGEVFRCHWDLDGALDNPNDKQEVGDGPVPWCSPDQLSFVAEQVSHHPPISAFYAEHVNKRIQFEAWVWTKSKFLGLSIGVHNIGKGTVTLIDLGEEYTLTFPNGYGRSILTVPWIELGGSVVIECVQTGHKANIEFLTKPFYGGKKHRVTCEIFAGAEKKPYYTAQGEWNTRMDGRWTDSGRSEILFEVSNMKPRRKRVAPVSRQLPHESRRVWRHVTCALRAADCDAATLAKRHVEQAQRDAVKHREASGEKWQTQLFSPRIEEGWEYKSPLKNRNVPCKSPQHKHSENDNTR
Summary
Similarity
Belongs to the OSBP family.
Uniprot
A0A2A4JRR8
A0A2A4JQI1
A0A2W1BK17
A0A3S2TL84
A0A194QCP8
H9J3T9
+ More
A0A2H1W479 A0A212F6R1 A0A212FAD1 A0A194R3L8 A0A0N1IPP5 A0A067RBW7 A0A2J7PVR3 A0A2J7PVR0 U5EPL9 A0A2J7PVR8 E1ZY54 A0A2A3E639 V9ILL0 A0A026VWB2 A0A088AUS5 A0A3L8DVI1 E2C3X1 A0A0L7QL07 A0A0J9RA69 A0A0B4KEI3 A0A182FHH4 A0A0Q5WAF7 A0A182RN30 A0A154PJN7 A0A182UNM1 A0A0R1DMD8 A0A2M4ANJ2 A0A182TFG8 A0A034VJP9 A0A182Y0Y2 A0A182LJI8 A0A1W4UZI8 A0A0K8V7G1 A0A0P8XPS7 N6V877 A0A182WKB7 A0A182GW92 A0A0K8VRR8 A0A182PM29 A0A195C6B8 A0A023EU46 A0A0Q9X8X0 A0A0A1X6H2 A0A151WF66 A0A0A1XSZ4 F4X8J4 A0A195FD32 A0A182NM09 W8B809 A0A195BGI4 W8AWZ9 A0A182MUJ5 K7J5X6 Q16NJ5 A0A158NBL4 A0A0L0BTX8 A0A232EUM3 A0A1L8DLI8 B4MQE7 A0A2M4CRK2 W8AT30 A0A2M4AJI6 A0A0J9R980 A0A0K8W677 A0A0Q9X828 A0A1L8DM07 A0A182QKQ0 A0A182J036 A0A2M4AJH9 A0A2C9GPJ3 A0A034VNZ2 A0A1I8NGF5 T1PKE8 A1Z814 A0A139WEQ1 A0A139WEI8 B3N6U0 W5JSM6 A0A195DS54 A0A1L8DMF2 B4NX11 A0A1W4UZ83 A0A1B6D6J0 A0A0A1WYD5 A0A182SBL0 B0VYY8 A0A0K8VAR0 Q28YZ8 A0A0Q9W5B5 A0A069DSR5 D3DMG7 B4KQ98 A0A0M4EFC4 B3MBK9
A0A2H1W479 A0A212F6R1 A0A212FAD1 A0A194R3L8 A0A0N1IPP5 A0A067RBW7 A0A2J7PVR3 A0A2J7PVR0 U5EPL9 A0A2J7PVR8 E1ZY54 A0A2A3E639 V9ILL0 A0A026VWB2 A0A088AUS5 A0A3L8DVI1 E2C3X1 A0A0L7QL07 A0A0J9RA69 A0A0B4KEI3 A0A182FHH4 A0A0Q5WAF7 A0A182RN30 A0A154PJN7 A0A182UNM1 A0A0R1DMD8 A0A2M4ANJ2 A0A182TFG8 A0A034VJP9 A0A182Y0Y2 A0A182LJI8 A0A1W4UZI8 A0A0K8V7G1 A0A0P8XPS7 N6V877 A0A182WKB7 A0A182GW92 A0A0K8VRR8 A0A182PM29 A0A195C6B8 A0A023EU46 A0A0Q9X8X0 A0A0A1X6H2 A0A151WF66 A0A0A1XSZ4 F4X8J4 A0A195FD32 A0A182NM09 W8B809 A0A195BGI4 W8AWZ9 A0A182MUJ5 K7J5X6 Q16NJ5 A0A158NBL4 A0A0L0BTX8 A0A232EUM3 A0A1L8DLI8 B4MQE7 A0A2M4CRK2 W8AT30 A0A2M4AJI6 A0A0J9R980 A0A0K8W677 A0A0Q9X828 A0A1L8DM07 A0A182QKQ0 A0A182J036 A0A2M4AJH9 A0A2C9GPJ3 A0A034VNZ2 A0A1I8NGF5 T1PKE8 A1Z814 A0A139WEQ1 A0A139WEI8 B3N6U0 W5JSM6 A0A195DS54 A0A1L8DMF2 B4NX11 A0A1W4UZ83 A0A1B6D6J0 A0A0A1WYD5 A0A182SBL0 B0VYY8 A0A0K8VAR0 Q28YZ8 A0A0Q9W5B5 A0A069DSR5 D3DMG7 B4KQ98 A0A0M4EFC4 B3MBK9
Pubmed
28756777
26354079
19121390
22118469
24845553
20798317
+ More
24508170 30249741 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17550304 25348373 25244985 20966253 15632085 26483478 24945155 25830018 21719571 24495485 20075255 17510324 21347285 26108605 28648823 25315136 26109357 26109356 18362917 19820115 20920257 23761445 26334808
24508170 30249741 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17550304 25348373 25244985 20966253 15632085 26483478 24945155 25830018 21719571 24495485 20075255 17510324 21347285 26108605 28648823 25315136 26109357 26109356 18362917 19820115 20920257 23761445 26334808
EMBL
NWSH01000769
PCG74314.1
PCG74315.1
KZ150130
PZC73070.1
RSAL01000070
+ More
RVE49171.1 KQ459185 KPJ03194.1 BABH01028800 BABH01028801 ODYU01006198 SOQ47837.1 AGBW02009986 OWR49399.1 AGBW02009493 OWR50691.1 KQ460949 KPJ10436.1 KQ460369 KPJ15428.1 KK852607 KDR20354.1 NEVH01020943 PNF20425.1 PNF20427.1 GANO01000122 JAB59749.1 PNF20426.1 GL435171 EFN73903.1 KZ288356 PBC27207.1 JR050866 AEY61371.1 KK107785 EZA47806.1 QOIP01000004 RLU23939.1 GL452364 EFN77357.1 KQ414934 KOC59307.1 CM002911 KMY92604.1 AE013599 AGB93371.1 CH954177 KQS70324.1 KQ434926 KZC11704.1 CM000157 KRJ98539.1 GGFK01009028 MBW42349.1 GAKP01015421 JAC43531.1 GDHF01017445 GDHF01004574 JAI34869.1 JAI47740.1 CH902619 KPU76627.1 CM000071 ENO01691.2 JXUM01019673 KQ560551 KXJ81924.1 GDHF01024779 GDHF01019092 GDHF01011074 JAI27535.1 JAI33222.1 JAI41240.1 KQ978219 KYM96409.1 GAPW01000685 JAC12913.1 CH933808 KRG04533.1 GBXI01007560 JAD06732.1 KQ983227 KYQ46467.1 GBXI01000609 JAD13683.1 GL888932 EGI57263.1 KQ981673 KYN38291.1 GAMC01017200 GAMC01017199 JAB89356.1 KQ976500 KYM83260.1 GAMC01017197 JAB89358.1 AXCM01001148 CH477822 EAT35922.1 ADTU01011210 JRES01001351 KNC23433.1 NNAY01002114 OXU22041.1 GFDF01006756 JAV07328.1 CH963849 EDW74336.1 GGFL01003755 MBW67933.1 GAMC01017201 GAMC01017198 JAB89357.1 GGFK01007632 MBW40953.1 KMY92606.1 GDHF01005683 JAI46631.1 KRG04534.1 GFDF01006714 JAV07370.1 AXCN02002331 GGFK01007624 MBW40945.1 APCN01004294 APCN01004295 APCN01004296 APCN01004297 GAKP01015422 JAC43530.1 KA648550 AFP63179.1 BT133169 AAF58878.4 AEZ68797.1 KQ971354 KYB26369.1 KYB26370.1 EDV58189.2 ADMH02000483 ETN66308.1 KQ980581 KYN15349.1 GFDF01006446 JAV07638.1 EDW89572.1 GEDC01016032 JAS21266.1 GBXI01010480 JAD03812.1 DS231813 EDS25733.1 GDHF01025426 GDHF01024825 GDHF01017934 GDHF01016388 GDHF01012166 GDHF01011657 JAI26888.1 JAI27489.1 JAI34380.1 JAI35926.1 JAI40148.1 JAI40657.1 EAL25815.4 CH940648 KRF80297.1 GBGD01001751 JAC87138.1 BT120036 ADA53575.1 EDW09226.2 CP012524 ALC40872.1 EDV37140.2
RVE49171.1 KQ459185 KPJ03194.1 BABH01028800 BABH01028801 ODYU01006198 SOQ47837.1 AGBW02009986 OWR49399.1 AGBW02009493 OWR50691.1 KQ460949 KPJ10436.1 KQ460369 KPJ15428.1 KK852607 KDR20354.1 NEVH01020943 PNF20425.1 PNF20427.1 GANO01000122 JAB59749.1 PNF20426.1 GL435171 EFN73903.1 KZ288356 PBC27207.1 JR050866 AEY61371.1 KK107785 EZA47806.1 QOIP01000004 RLU23939.1 GL452364 EFN77357.1 KQ414934 KOC59307.1 CM002911 KMY92604.1 AE013599 AGB93371.1 CH954177 KQS70324.1 KQ434926 KZC11704.1 CM000157 KRJ98539.1 GGFK01009028 MBW42349.1 GAKP01015421 JAC43531.1 GDHF01017445 GDHF01004574 JAI34869.1 JAI47740.1 CH902619 KPU76627.1 CM000071 ENO01691.2 JXUM01019673 KQ560551 KXJ81924.1 GDHF01024779 GDHF01019092 GDHF01011074 JAI27535.1 JAI33222.1 JAI41240.1 KQ978219 KYM96409.1 GAPW01000685 JAC12913.1 CH933808 KRG04533.1 GBXI01007560 JAD06732.1 KQ983227 KYQ46467.1 GBXI01000609 JAD13683.1 GL888932 EGI57263.1 KQ981673 KYN38291.1 GAMC01017200 GAMC01017199 JAB89356.1 KQ976500 KYM83260.1 GAMC01017197 JAB89358.1 AXCM01001148 CH477822 EAT35922.1 ADTU01011210 JRES01001351 KNC23433.1 NNAY01002114 OXU22041.1 GFDF01006756 JAV07328.1 CH963849 EDW74336.1 GGFL01003755 MBW67933.1 GAMC01017201 GAMC01017198 JAB89357.1 GGFK01007632 MBW40953.1 KMY92606.1 GDHF01005683 JAI46631.1 KRG04534.1 GFDF01006714 JAV07370.1 AXCN02002331 GGFK01007624 MBW40945.1 APCN01004294 APCN01004295 APCN01004296 APCN01004297 GAKP01015422 JAC43530.1 KA648550 AFP63179.1 BT133169 AAF58878.4 AEZ68797.1 KQ971354 KYB26369.1 KYB26370.1 EDV58189.2 ADMH02000483 ETN66308.1 KQ980581 KYN15349.1 GFDF01006446 JAV07638.1 EDW89572.1 GEDC01016032 JAS21266.1 GBXI01010480 JAD03812.1 DS231813 EDS25733.1 GDHF01025426 GDHF01024825 GDHF01017934 GDHF01016388 GDHF01012166 GDHF01011657 JAI26888.1 JAI27489.1 JAI34380.1 JAI35926.1 JAI40148.1 JAI40657.1 EAL25815.4 CH940648 KRF80297.1 GBGD01001751 JAC87138.1 BT120036 ADA53575.1 EDW09226.2 CP012524 ALC40872.1 EDV37140.2
Proteomes
UP000218220
UP000283053
UP000053268
UP000005204
UP000007151
UP000053240
+ More
UP000027135 UP000235965 UP000000311 UP000242457 UP000053097 UP000005203 UP000279307 UP000008237 UP000053825 UP000000803 UP000069272 UP000008711 UP000075900 UP000076502 UP000075903 UP000002282 UP000075902 UP000076408 UP000075882 UP000192221 UP000007801 UP000001819 UP000075920 UP000069940 UP000249989 UP000075885 UP000078542 UP000009192 UP000075809 UP000007755 UP000078541 UP000075884 UP000078540 UP000075883 UP000002358 UP000008820 UP000005205 UP000037069 UP000215335 UP000007798 UP000075886 UP000075880 UP000075840 UP000095301 UP000007266 UP000000673 UP000078492 UP000075901 UP000002320 UP000008792 UP000092553
UP000027135 UP000235965 UP000000311 UP000242457 UP000053097 UP000005203 UP000279307 UP000008237 UP000053825 UP000000803 UP000069272 UP000008711 UP000075900 UP000076502 UP000075903 UP000002282 UP000075902 UP000076408 UP000075882 UP000192221 UP000007801 UP000001819 UP000075920 UP000069940 UP000249989 UP000075885 UP000078542 UP000009192 UP000075809 UP000007755 UP000078541 UP000075884 UP000078540 UP000075883 UP000002358 UP000008820 UP000005205 UP000037069 UP000215335 UP000007798 UP000075886 UP000075880 UP000075840 UP000095301 UP000007266 UP000000673 UP000078492 UP000075901 UP000002320 UP000008792 UP000092553
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JRR8
A0A2A4JQI1
A0A2W1BK17
A0A3S2TL84
A0A194QCP8
H9J3T9
+ More
A0A2H1W479 A0A212F6R1 A0A212FAD1 A0A194R3L8 A0A0N1IPP5 A0A067RBW7 A0A2J7PVR3 A0A2J7PVR0 U5EPL9 A0A2J7PVR8 E1ZY54 A0A2A3E639 V9ILL0 A0A026VWB2 A0A088AUS5 A0A3L8DVI1 E2C3X1 A0A0L7QL07 A0A0J9RA69 A0A0B4KEI3 A0A182FHH4 A0A0Q5WAF7 A0A182RN30 A0A154PJN7 A0A182UNM1 A0A0R1DMD8 A0A2M4ANJ2 A0A182TFG8 A0A034VJP9 A0A182Y0Y2 A0A182LJI8 A0A1W4UZI8 A0A0K8V7G1 A0A0P8XPS7 N6V877 A0A182WKB7 A0A182GW92 A0A0K8VRR8 A0A182PM29 A0A195C6B8 A0A023EU46 A0A0Q9X8X0 A0A0A1X6H2 A0A151WF66 A0A0A1XSZ4 F4X8J4 A0A195FD32 A0A182NM09 W8B809 A0A195BGI4 W8AWZ9 A0A182MUJ5 K7J5X6 Q16NJ5 A0A158NBL4 A0A0L0BTX8 A0A232EUM3 A0A1L8DLI8 B4MQE7 A0A2M4CRK2 W8AT30 A0A2M4AJI6 A0A0J9R980 A0A0K8W677 A0A0Q9X828 A0A1L8DM07 A0A182QKQ0 A0A182J036 A0A2M4AJH9 A0A2C9GPJ3 A0A034VNZ2 A0A1I8NGF5 T1PKE8 A1Z814 A0A139WEQ1 A0A139WEI8 B3N6U0 W5JSM6 A0A195DS54 A0A1L8DMF2 B4NX11 A0A1W4UZ83 A0A1B6D6J0 A0A0A1WYD5 A0A182SBL0 B0VYY8 A0A0K8VAR0 Q28YZ8 A0A0Q9W5B5 A0A069DSR5 D3DMG7 B4KQ98 A0A0M4EFC4 B3MBK9
A0A2H1W479 A0A212F6R1 A0A212FAD1 A0A194R3L8 A0A0N1IPP5 A0A067RBW7 A0A2J7PVR3 A0A2J7PVR0 U5EPL9 A0A2J7PVR8 E1ZY54 A0A2A3E639 V9ILL0 A0A026VWB2 A0A088AUS5 A0A3L8DVI1 E2C3X1 A0A0L7QL07 A0A0J9RA69 A0A0B4KEI3 A0A182FHH4 A0A0Q5WAF7 A0A182RN30 A0A154PJN7 A0A182UNM1 A0A0R1DMD8 A0A2M4ANJ2 A0A182TFG8 A0A034VJP9 A0A182Y0Y2 A0A182LJI8 A0A1W4UZI8 A0A0K8V7G1 A0A0P8XPS7 N6V877 A0A182WKB7 A0A182GW92 A0A0K8VRR8 A0A182PM29 A0A195C6B8 A0A023EU46 A0A0Q9X8X0 A0A0A1X6H2 A0A151WF66 A0A0A1XSZ4 F4X8J4 A0A195FD32 A0A182NM09 W8B809 A0A195BGI4 W8AWZ9 A0A182MUJ5 K7J5X6 Q16NJ5 A0A158NBL4 A0A0L0BTX8 A0A232EUM3 A0A1L8DLI8 B4MQE7 A0A2M4CRK2 W8AT30 A0A2M4AJI6 A0A0J9R980 A0A0K8W677 A0A0Q9X828 A0A1L8DM07 A0A182QKQ0 A0A182J036 A0A2M4AJH9 A0A2C9GPJ3 A0A034VNZ2 A0A1I8NGF5 T1PKE8 A1Z814 A0A139WEQ1 A0A139WEI8 B3N6U0 W5JSM6 A0A195DS54 A0A1L8DMF2 B4NX11 A0A1W4UZ83 A0A1B6D6J0 A0A0A1WYD5 A0A182SBL0 B0VYY8 A0A0K8VAR0 Q28YZ8 A0A0Q9W5B5 A0A069DSR5 D3DMG7 B4KQ98 A0A0M4EFC4 B3MBK9
PDB
4PH7
E-value=4.63002e-41,
Score=422
Ontologies
PANTHER
Topology
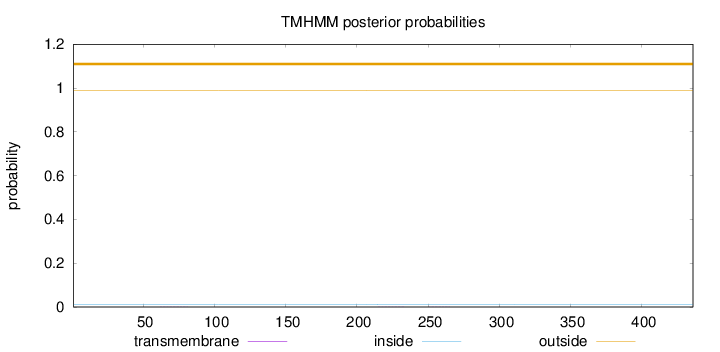
Length:
436
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01113
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.01245
outside
1 - 436
Population Genetic Test Statistics
Pi
23.312438
Theta
20.147295
Tajima's D
0.39158
CLR
1.162439
CSRT
0.493125343732813
Interpretation
Uncertain
