Gene
KWMTBOMO11712
Pre Gene Modal
BGIBMGA004170
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.369 Nuclear Reliability : 1.832
Sequence
CDS
ATGGAGTGGCTCCGAAAATATTACCACACAGTTTTCTACGATAGAGCGGATCAGAGGGTCAAAGATTGGCTGTTCATGACGTCACCATGGCCAATCGTGGCGATCCTAACGACTTATCTGCTGCTCATCAAGTTTATTCTGCCCATCTACATGAGGAACAAGAGACCTTACAGTTTGAAAGGCGTTATAAAAAGCTACAACGTGACTATGATTGTTTTCAACTTGGTCGTTGCTTGGGGGATAGCAACGTCAGGTTGGACAACCACATACCATTTCGGATGCGTTCTACCAGATTATACCACGAAACCGGAACCAATGAGGATGCTCACTTTCATGTGGTGGACATTGACTATCAAGATGACAGAGCTATTGGAGACCGTAATATTTTTGTTGAGAAAAAAAAATCAACAGGCCTCGTTCCTTCACGTGTACCATCACGTCATCTCTTTGGTTATGATATGGTGTGGTGTCAAATATATTGGAGGAGGAGACCAACGATTGGAAAGATATGGTGTGGGAATTCTTCTCACAAAGGAAATTTCCAAATCAGTCATAAATTTTGTTCCCCTCTCAAACCGGGCCATGTTGATTAAATTAGAAGCCAAACCATTTAACATCAACATTGTCCAAGTTTATGCGCCTACAGGGGATAAACCAATGGGCCAGATAGAGGACTTATATAGGGCTATTGATACCTTATTAAAATCTACTAAGAATCACGAGTTAGTTATCGTTATGGGTGACATGAATGCGAAGGTGGGTTCGACCATTACTCCTGGAGTGACTGGTGCTTATGGCCTTGGAAACAGAAACGATAGAGGTGATACGCTTATTGAGTTTTGCCAGGATAATAACCTCATAATTGCGAACACCTTCTTTAAGCTACCACCACGTCGGCTATACACATGGACATCACCAATGCACACAGTAGACCGCGTAGTCAGAAACCAAATTGACTATGTTATGATCAACAGAAGGTATAGAAATTGTATCAAGAAGGCGCAAACCTACCCCGGTGCAGACATTGGGTCAGATCATAACCCAGTGGTGGTAGACGTAGCATGCAAGCTGAAGAAGTTGCAGCCAAAGAGAATGGATGGAAGGCCAAACGTACGGAAGCTTGCTGAGCCCGACACACGAGCGAAGACCCACACCGCTATTAATCAATGGGCCAACGATCGTCGCACTGAAACCACTGCCTCTGACCTCGACACGTGGACTAGTCTAAAGCGAAGTGTAAAAGATATATGTCACCAATACCTCCAACCATCACAACTGGTCAAAAAACAAGATTGGATGACAGATTCAATTTTGCAGCTTATGGAAGACAGGAGGCAATTTAAGGGTACAAATCCAGTAATGTATAAGCAAATCGATGTCACCATTCGAAAAGAGATTGCGAAAGCCAAGAATGATTACTACAGCAATAAGTGCGCGCACTTGGAAGATCTCCTTCGTAAACATGACAGCTTCAATGCGCACAAGCACATCAAGGAAATGGTTGGTCATAGACGGAAATCAATGAATACTCTGACCGATGCAGGTGGGAACCTCATTCTTGACACCGATGGCAAAAAGAGAGTGTGGTGTGACTATGTTGCAGAGATGTTTGCGGACTCATCGAGGAACATTTCACCGATCACTGATCGGACTGGTCCAGCTATGTTGCGATCTGAGGTTTTGCGGGCCTTGAAAGCTGCTAAAACAGGCAATCTTCCTAGTGACTGGCTAAAATCCACATTCATCACGATACCCAAGAAGCAAAATGCCAAAAAATGTGGAGAGTATCGTATGATTAGTCTGATGAGCCATGTCCTAAAAGTATTCTTGAACATCATACAGAATAGAATACGGCCAAAATGTGATGAACAACTGGGCGATAGCCAATTTGGATTTCGATCTGGTGTTGGCACCAGAGAAGCTCTTTTTGCTCTCAACGTTCTCGTACAGAAATGCAGAGACATGCAAACTGATGTCTTTTTGTGCTTCATAGACTACGAAAAGGCATTTGACCGAGTTAAACATCATCAACTTTTTAGCCTTTTATGTGACATTGGTCTAGATGGTAAGGACGTCAGAATCATCCGCAACCTGTACGAGAAGCAGGTGGCTACGATAAGAGTCGAAAATGAGGAAACCGACCAAGTTGAGATCTGTCGTGGGGTCCGGCAGGGGTGTGTCTTATCCCCGTTGCTTTTTAACATCTACTCAGAGGCTGTGATGTCAAAAGCTCTTGAAAATCTTGAAGTGGGAATAGGAATCAACGGCAGGGTCGTTAACAACTTGCGTTATGCGGACGACACTATCCTTATTGCAGCATCGGAAGCCGATTTGCAAGCAATTGTGAATAAGGTTAATGAGTGCAGCGAAGAAGCCGGGTTGTCGATAAACATAAGCAAGACCAAGTTCATGGTAGTCTCAAGAAACCCGGATTTGAGTTCAAGCGTATCTGTAGCTGGGAAACAACTAGAGCGGGTACGACAGTACAAGTATTTAGGGGCTTGGGTAAACGAAGCATGGGAATCTGATCAAGAAATCAAGACCCGGATAGAAACTGCTCGAGCCTCTTTTAATAAAATGAGGAAAGTTCTTTGCTGTCGTCAACTGAATATAAAGCTCCGAGTCAGGCTACTCATGTGCTATATCTGGCCTATTGTCATGTATGGTTGCGAAACGTGGACCCTGAAAGAGGACACTACAAGACGCCTACAGGCATTCGAGATGTGGTGCTACCGTCGCATGTTACGCATTGGTTGGACGCAGAGGGTCAGGAACGAAACCGTGCTTCAGCGCGTTCATATGTCCCGGAAAATGCTGCCTGCTATCAAAAAGCGCAAGATAGAGTATCTCGGCCACGTGCTCAGGAACGATCGATACATGCTGCTCCAGCTGATAATTATGGGCAAAGTGGACGGAAAGAGACGTGCTGGGCGAAGAAAGAAATCATGGCTCCGGAATATCCGGGAGTGGACCGGTATTGCCTCCGTCGAACAGCTGTTCCGGCTGGCTGCAGATAAAAACGAATATAGAAAGCTAACGGCCAACCTTCAGGATTGA
Protein
MEWLRKYYHTVFYDRADQRVKDWLFMTSPWPIVAILTTYLLLIKFILPIYMRNKRPYSLKGVIKSYNVTMIVFNLVVAWGIATSGWTTTYHFGCVLPDYTTKPEPMRMLTFMWWTLTIKMTELLETVIFLLRKKNQQASFLHVYHHVISLVMIWCGVKYIGGGDQRLERYGVGILLTKEISKSVINFVPLSNRAMLIKLEAKPFNINIVQVYAPTGDKPMGQIEDLYRAIDTLLKSTKNHELVIVMGDMNAKVGSTITPGVTGAYGLGNRNDRGDTLIEFCQDNNLIIANTFFKLPPRRLYTWTSPMHTVDRVVRNQIDYVMINRRYRNCIKKAQTYPGADIGSDHNPVVVDVACKLKKLQPKRMDGRPNVRKLAEPDTRAKTHTAINQWANDRRTETTASDLDTWTSLKRSVKDICHQYLQPSQLVKKQDWMTDSILQLMEDRRQFKGTNPVMYKQIDVTIRKEIAKAKNDYYSNKCAHLEDLLRKHDSFNAHKHIKEMVGHRRKSMNTLTDAGGNLILDTDGKKRVWCDYVAEMFADSSRNISPITDRTGPAMLRSEVLRALKAAKTGNLPSDWLKSTFITIPKKQNAKKCGEYRMISLMSHVLKVFLNIIQNRIRPKCDEQLGDSQFGFRSGVGTREALFALNVLVQKCRDMQTDVFLCFIDYEKAFDRVKHHQLFSLLCDIGLDGKDVRIIRNLYEKQVATIRVENEETDQVEICRGVRQGCVLSPLLFNIYSEAVMSKALENLEVGIGINGRVVNNLRYADDTILIAASEADLQAIVNKVNECSEEAGLSINISKTKFMVVSRNPDLSSSVSVAGKQLERVRQYKYLGAWVNEAWESDQEIKTRIETARASFNKMRKVLCCRQLNIKLRVRLLMCYIWPIVMYGCETWTLKEDTTRRLQAFEMWCYRRMLRIGWTQRVRNETVLQRVHMSRKMLPAIKKRKIEYLGHVLRNDRYMLLQLIIMGKVDGKRRAGRRKKSWLRNIREWTGIASVEQLFRLAADKNEYRKLTANLQD
Summary
Uniprot
D5LB38
A0A2A4JBK9
A0A1W7RAL1
A0A2V9X8L4
A0A2S2PMS6
A0A076N1B6
+ More
A0A3S3P648 A0A2J7RP47 A0A2J7Q6A8 A0A2J7Q5D5 A0A2J7QI56 A0A2J7PTY0 A0A2J7PW81 A0A2J7QR01 A0A2J7REE7 A0A2J7PMJ3 A0A2J7PQA3 A0A2J7RLI7 D7F162 A0A2J7QEJ0 A0A2J7Q6P7 A0A2J7QGT4 A0A2J7Q2Z3 A0A2J7RGX2 A0A2J7QKB0 A0A2J7Q0C6 A0A2J7QH51 A0A2J7QUN7 A0A2J7R4R4 A0A2J7QGJ2 A0A2J7PHM9 A0A2J7Q3T8 A0A2J7QGX8 A0A146LUY8 D7F163 A0A076N3Z4 A0A2A4JXP9 D7F168 D7F161 D5LB39 A0A2J7QCG2 A0A2J7QJJ9 A0A2J7PT27 A0A2J7QUD0 A0A2J7R8G1 A0A2J7RIL8 A0A2J7R8P5 A0A2J7R901 A0A2J7QMC5 A0A2J7PX03 W5MYT3 A0A2J7PBF1 A0A2J7PF68 A0A2J7RPX5 A0A2J7Q649 A0A2J7RBL1 A0A2J7QSZ5 A0A2J7PVW0 A0A2J7PNH4 A0A2J7PSH5 A0A2J7PC89 A0A2J7QNQ3 A0A2J7Q7C6 A0A2J7RRE4 A0A2J7RBL5 A0A2J7PZN7 W5NGM3 A0A2J7Q6T5 A0A2J7Q3K9 A0A2J7PFC5 A0A2W1BNH4 W4Z133 A0A2J7QLQ1
A0A3S3P648 A0A2J7RP47 A0A2J7Q6A8 A0A2J7Q5D5 A0A2J7QI56 A0A2J7PTY0 A0A2J7PW81 A0A2J7QR01 A0A2J7REE7 A0A2J7PMJ3 A0A2J7PQA3 A0A2J7RLI7 D7F162 A0A2J7QEJ0 A0A2J7Q6P7 A0A2J7QGT4 A0A2J7Q2Z3 A0A2J7RGX2 A0A2J7QKB0 A0A2J7Q0C6 A0A2J7QH51 A0A2J7QUN7 A0A2J7R4R4 A0A2J7QGJ2 A0A2J7PHM9 A0A2J7Q3T8 A0A2J7QGX8 A0A146LUY8 D7F163 A0A076N3Z4 A0A2A4JXP9 D7F168 D7F161 D5LB39 A0A2J7QCG2 A0A2J7QJJ9 A0A2J7PT27 A0A2J7QUD0 A0A2J7R8G1 A0A2J7RIL8 A0A2J7R8P5 A0A2J7R901 A0A2J7QMC5 A0A2J7PX03 W5MYT3 A0A2J7PBF1 A0A2J7PF68 A0A2J7RPX5 A0A2J7Q649 A0A2J7RBL1 A0A2J7QSZ5 A0A2J7PVW0 A0A2J7PNH4 A0A2J7PSH5 A0A2J7PC89 A0A2J7QNQ3 A0A2J7Q7C6 A0A2J7RRE4 A0A2J7RBL5 A0A2J7PZN7 W5NGM3 A0A2J7Q6T5 A0A2J7Q3K9 A0A2J7PFC5 A0A2W1BNH4 W4Z133 A0A2J7QLQ1
EMBL
GU815089
ADF18552.1
NWSH01001998
PCG69477.1
GFAH01000222
JAV48167.1
+ More
QIAP01000240 PYX61602.1 GGMR01018096 MBY30715.1 KF881087 AIJ27486.1 NCKU01004395 RWS06003.1 NEVH01002141 PNF42599.1 NEVH01017534 PNF24114.1 NEVH01017570 PNF23798.1 NEVH01013959 PNF28275.1 NEVH01021219 PNF19787.1 NEVH01020937 PNF20581.1 NEVH01012082 PNF31006.1 NEVH01005277 PNF39212.1 NEVH01023985 PNF17546.1 NEVH01022640 NEVH01008218 NEVH01005885 PNF18513.1 PNF34430.1 PNF38481.1 NEVH01002684 PNF41704.1 FJ265547 ADI61815.1 NEVH01015309 PNF27009.1 NEVH01017460 PNF24248.1 NEVH01014359 PNF27789.1 NEVH01019075 PNF22955.1 NEVH01003750 PNF40091.1 NEVH01013275 PNF29009.1 NEVH01019971 PNF22037.1 NEVH01014358 PNF27914.1 NEVH01023960 NEVH01010501 PNF17732.1 PNF32290.1 NEVH01007402 PNF35815.1 NEVH01018372 NEVH01014371 NEVH01011208 NEVH01002697 NEVH01002552 PNF23678.1 PNF27673.1 PNF31697.1 PNF41592.1 PNF42118.1 NEVH01025135 PNF15828.1 NEVH01019064 PNF23230.1 PNF27813.1 GDHC01007098 GDHC01001590 JAQ11531.1 JAQ17039.1 FJ265548 ADI61816.1 KF881086 AIJ27485.1 NWSH01000469 PCG76200.1 FJ265553 ADI61821.1 FJ265546 ADI61814.1 GU815090 ADF18553.1 NEVH01016289 PNF26277.1 NEVH01013553 PNF28765.1 NEVH01021921 PNF19488.1 NEVH01011186 PNF32183.1 NEVH01016335 NEVH01006721 PNF25462.1 PNF37115.1 NEVH01003500 PNF40666.1 PNF37211.1 PNF37310.1 NEVH01013215 PNF29703.1 NEVH01020865 PNF20863.1 AHAT01017643 NEVH01027104 PNF13657.1 NEVH01026085 PNF14968.1 NEVH01020851 NEVH01013954 NEVH01006574 NEVH01001358 NEVH01001354 PNF21237.1 PNF28332.1 PNF37782.1 PNF42792.1 PNF42884.1 NEVH01017540 PNF24056.1 NEVH01005896 PNF38208.1 NEVH01011205 PNF31711.1 NEVH01020940 PNF20478.1 NEVH01023953 PNF17877.1 NEVH01021931 PNF19256.1 NEVH01027067 PNF13944.1 NEVH01012564 PNF30212.1 NEVH01017443 PNF24485.1 NEVH01000609 PNF43402.1 PNF38212.1 NEVH01020333 PNF21790.1 AHAT01008485 NEVH01017452 PNF24293.1 NEVH01019066 PNF23173.1 NEVH01025668 PNF15041.1 KZ150025 PZC74827.1 AAGJ04171899 NEVH01013243 PNF29503.1
QIAP01000240 PYX61602.1 GGMR01018096 MBY30715.1 KF881087 AIJ27486.1 NCKU01004395 RWS06003.1 NEVH01002141 PNF42599.1 NEVH01017534 PNF24114.1 NEVH01017570 PNF23798.1 NEVH01013959 PNF28275.1 NEVH01021219 PNF19787.1 NEVH01020937 PNF20581.1 NEVH01012082 PNF31006.1 NEVH01005277 PNF39212.1 NEVH01023985 PNF17546.1 NEVH01022640 NEVH01008218 NEVH01005885 PNF18513.1 PNF34430.1 PNF38481.1 NEVH01002684 PNF41704.1 FJ265547 ADI61815.1 NEVH01015309 PNF27009.1 NEVH01017460 PNF24248.1 NEVH01014359 PNF27789.1 NEVH01019075 PNF22955.1 NEVH01003750 PNF40091.1 NEVH01013275 PNF29009.1 NEVH01019971 PNF22037.1 NEVH01014358 PNF27914.1 NEVH01023960 NEVH01010501 PNF17732.1 PNF32290.1 NEVH01007402 PNF35815.1 NEVH01018372 NEVH01014371 NEVH01011208 NEVH01002697 NEVH01002552 PNF23678.1 PNF27673.1 PNF31697.1 PNF41592.1 PNF42118.1 NEVH01025135 PNF15828.1 NEVH01019064 PNF23230.1 PNF27813.1 GDHC01007098 GDHC01001590 JAQ11531.1 JAQ17039.1 FJ265548 ADI61816.1 KF881086 AIJ27485.1 NWSH01000469 PCG76200.1 FJ265553 ADI61821.1 FJ265546 ADI61814.1 GU815090 ADF18553.1 NEVH01016289 PNF26277.1 NEVH01013553 PNF28765.1 NEVH01021921 PNF19488.1 NEVH01011186 PNF32183.1 NEVH01016335 NEVH01006721 PNF25462.1 PNF37115.1 NEVH01003500 PNF40666.1 PNF37211.1 PNF37310.1 NEVH01013215 PNF29703.1 NEVH01020865 PNF20863.1 AHAT01017643 NEVH01027104 PNF13657.1 NEVH01026085 PNF14968.1 NEVH01020851 NEVH01013954 NEVH01006574 NEVH01001358 NEVH01001354 PNF21237.1 PNF28332.1 PNF37782.1 PNF42792.1 PNF42884.1 NEVH01017540 PNF24056.1 NEVH01005896 PNF38208.1 NEVH01011205 PNF31711.1 NEVH01020940 PNF20478.1 NEVH01023953 PNF17877.1 NEVH01021931 PNF19256.1 NEVH01027067 PNF13944.1 NEVH01012564 PNF30212.1 NEVH01017443 PNF24485.1 NEVH01000609 PNF43402.1 PNF38212.1 NEVH01020333 PNF21790.1 AHAT01008485 NEVH01017452 PNF24293.1 NEVH01019066 PNF23173.1 NEVH01025668 PNF15041.1 KZ150025 PZC74827.1 AAGJ04171899 NEVH01013243 PNF29503.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D5LB38
A0A2A4JBK9
A0A1W7RAL1
A0A2V9X8L4
A0A2S2PMS6
A0A076N1B6
+ More
A0A3S3P648 A0A2J7RP47 A0A2J7Q6A8 A0A2J7Q5D5 A0A2J7QI56 A0A2J7PTY0 A0A2J7PW81 A0A2J7QR01 A0A2J7REE7 A0A2J7PMJ3 A0A2J7PQA3 A0A2J7RLI7 D7F162 A0A2J7QEJ0 A0A2J7Q6P7 A0A2J7QGT4 A0A2J7Q2Z3 A0A2J7RGX2 A0A2J7QKB0 A0A2J7Q0C6 A0A2J7QH51 A0A2J7QUN7 A0A2J7R4R4 A0A2J7QGJ2 A0A2J7PHM9 A0A2J7Q3T8 A0A2J7QGX8 A0A146LUY8 D7F163 A0A076N3Z4 A0A2A4JXP9 D7F168 D7F161 D5LB39 A0A2J7QCG2 A0A2J7QJJ9 A0A2J7PT27 A0A2J7QUD0 A0A2J7R8G1 A0A2J7RIL8 A0A2J7R8P5 A0A2J7R901 A0A2J7QMC5 A0A2J7PX03 W5MYT3 A0A2J7PBF1 A0A2J7PF68 A0A2J7RPX5 A0A2J7Q649 A0A2J7RBL1 A0A2J7QSZ5 A0A2J7PVW0 A0A2J7PNH4 A0A2J7PSH5 A0A2J7PC89 A0A2J7QNQ3 A0A2J7Q7C6 A0A2J7RRE4 A0A2J7RBL5 A0A2J7PZN7 W5NGM3 A0A2J7Q6T5 A0A2J7Q3K9 A0A2J7PFC5 A0A2W1BNH4 W4Z133 A0A2J7QLQ1
A0A3S3P648 A0A2J7RP47 A0A2J7Q6A8 A0A2J7Q5D5 A0A2J7QI56 A0A2J7PTY0 A0A2J7PW81 A0A2J7QR01 A0A2J7REE7 A0A2J7PMJ3 A0A2J7PQA3 A0A2J7RLI7 D7F162 A0A2J7QEJ0 A0A2J7Q6P7 A0A2J7QGT4 A0A2J7Q2Z3 A0A2J7RGX2 A0A2J7QKB0 A0A2J7Q0C6 A0A2J7QH51 A0A2J7QUN7 A0A2J7R4R4 A0A2J7QGJ2 A0A2J7PHM9 A0A2J7Q3T8 A0A2J7QGX8 A0A146LUY8 D7F163 A0A076N3Z4 A0A2A4JXP9 D7F168 D7F161 D5LB39 A0A2J7QCG2 A0A2J7QJJ9 A0A2J7PT27 A0A2J7QUD0 A0A2J7R8G1 A0A2J7RIL8 A0A2J7R8P5 A0A2J7R901 A0A2J7QMC5 A0A2J7PX03 W5MYT3 A0A2J7PBF1 A0A2J7PF68 A0A2J7RPX5 A0A2J7Q649 A0A2J7RBL1 A0A2J7QSZ5 A0A2J7PVW0 A0A2J7PNH4 A0A2J7PSH5 A0A2J7PC89 A0A2J7QNQ3 A0A2J7Q7C6 A0A2J7RRE4 A0A2J7RBL5 A0A2J7PZN7 W5NGM3 A0A2J7Q6T5 A0A2J7Q3K9 A0A2J7PFC5 A0A2W1BNH4 W4Z133 A0A2J7QLQ1
PDB
5HHL
E-value=4.11769e-12,
Score=176
Ontologies
PATHWAY
GO
Topology
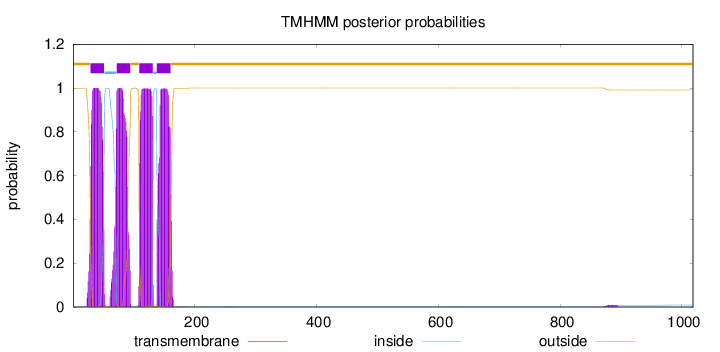
Length:
1018
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
87.4000500000001
Exp number, first 60 AAs:
21.88899
Total prob of N-in:
0.00073
POSSIBLE N-term signal
sequence
outside
1 - 28
TMhelix
29 - 51
inside
52 - 71
TMhelix
72 - 94
outside
95 - 108
TMhelix
109 - 131
inside
132 - 137
TMhelix
138 - 160
outside
161 - 1018
Population Genetic Test Statistics
Pi
25.070393
Theta
27.248279
Tajima's D
-0.62575
CLR
0.656864
CSRT
0.222388880555972
Interpretation
Uncertain
