Pre Gene Modal
BGIBMGA004172
Annotation
Leucine-rich_transmembrane_protein_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 1.848
Sequence
CDS
ATGCCGAAAACGAATTTGACATTATTGCAATATACATATAGGCTACAGGATTTAAATTTGACAAATAATCAAATTGAGAGAATCGCTGGCTCACCTTTTTACTACCTCGGCAAATTAGAAACCATTGATTTATCTCATAACAGGATTAGCGATCTGGAAGAACTATTTCGTTTTGAAACGCGACCATACAAACTCAACAAATTAGTACTTGCCAACAACAATATAGAAGAACTGCCCCAAGACGCTTTTGACGAACTGTCATCTCTCGTCGAACTCGATCTCTCATACAATCGCATTTCAAATCTGAACGAAGAGCCATTCGCCAATCTATCCAGTCTCGAAACACTAAGACTGAACAATAATTCCATCAAAAACCTAAACGGTGCAGTTAATAACCTTCAAAATCTGAAGCACCTATATTTAAGGGGAAACCAAATACAGCATATCGACGTAGAATCACTTAAAATCATAATACACCTGGAAACGTTCGACGTGTCGAGGAATCAACTCGAAAAAATCAACTCAGTCATGTTCTCGAGGCACTGGAAACACATGGGGGGCCATTCCATTTGTAAAATCATACTGTCCGAAAACCGCATAGCATCATTACCGAATGCATCAATGGAAATATCTGCTAGAAACGTCCGTGACTTGAGACGTTACAGCGTGGATGTGCTGACCGAATTAGATTTGTCGAACAATGAAATAACAGAAATAGCTTACAATGCGTTCCATTCTCTTTTGAAAATGATCTCATTGGATCTATCCCGGAACCAAATTACCGATTTTAGTGTAAACGCTGACGATCTCCAATTCGTAAAGAAACTGAACCTAAGCGGAAACCATATTTTCCAATTATATTACAAGAGCTTCTCGGCAATGGGAAACCTTGAGAATTTGGATCTCTCTCACAACAAACTTTACGAAATACCAGAAACAGCTTTTAAGAATTACTTTAGCCTTAGAATTCTGAATATGACGTTTAATGATATAGACAGCGTTGAGAATTTACGTATAACCAGATTCCATCCCGCTGGAGGTGTTCTGGACTTGTCAAACAACGGCTTGACGCAATTGAGGATACCATATGGTGAAGGAGCCCGTTTGATGATTTTGAGATTACACTCCAATAATATATCAGATGCTAAGCTAATCGAATTGATTCATCAGAACGATCTGAAAAATTTGGATCTCAGTCATAACAAGATACGCGAGCTTAATGACACATCTTTACGTATTCCCGTCAGTCTCAATTATTTAGATATCAGTTTTAACATGATCATTAGAATTGGACCATCATCTTTTTATCGTGCCGGTCATTTGAAAACTCTACGCTTGTCACACAATCAACTGACCAAGATTGAGCATGGAGCATTTCATGGATTATCAGCGCTTCTCAACCTTGATTTGTCTTACAACAAATTTGGGTACTTCAATTCTGAGTGGCTGATGGACCTTAAAAGTTTGGTTGTTCTTTCTCTCAGGAACAACTGGATGCATAATTTGGACTATAAGAGTTGGTTTGGCCACAAACCTGACCTGCGAGTGAATATCGAAGGAAATAGATTGTCTTGTGAGTGGCTTAGCGGTGTGTTGCAGAATTTTAATAACGGCTTCCTGAAGATGCACCCTGTCGTGTTGACGCCAACAATTTCAAGCCACTCTATAGAAGGCATTCCGTGTATCGAAAAAACTGAGGTCCTAAGTCAAATCTACAGCAGTCCGATGATGAGCGATGACCGTCTACTTGTGTCGAACCAGAAGATCCTGGAGGCTGTTAAGGAGCAGACGTATTATCTGCGCAGATACGTCATGCGTTCTCTTGTGGAGGAAACAGAAAGGGCGAAGCTGTAA
Protein
MPKTNLTLLQYTYRLQDLNLTNNQIERIAGSPFYYLGKLETIDLSHNRISDLEELFRFETRPYKLNKLVLANNNIEELPQDAFDELSSLVELDLSYNRISNLNEEPFANLSSLETLRLNNNSIKNLNGAVNNLQNLKHLYLRGNQIQHIDVESLKIIIHLETFDVSRNQLEKINSVMFSRHWKHMGGHSICKIILSENRIASLPNASMEISARNVRDLRRYSVDVLTELDLSNNEITEIAYNAFHSLLKMISLDLSRNQITDFSVNADDLQFVKKLNLSGNHIFQLYYKSFSAMGNLENLDLSHNKLYEIPETAFKNYFSLRILNMTFNDIDSVENLRITRFHPAGGVLDLSNNGLTQLRIPYGEGARLMILRLHSNNISDAKLIELIHQNDLKNLDLSHNKIRELNDTSLRIPVSLNYLDISFNMIIRIGPSSFYRAGHLKTLRLSHNQLTKIEHGAFHGLSALLNLDLSYNKFGYFNSEWLMDLKSLVVLSLRNNWMHNLDYKSWFGHKPDLRVNIEGNRLSCEWLSGVLQNFNNGFLKMHPVVLTPTISSHSIEGIPCIEKTEVLSQIYSSPMMSDDRLLVSNQKILEAVKEQTYYLRRYVMRSLVEETERAKL
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Belongs to the Toll-like receptor family.
Belongs to the Toll-like receptor family.
Uniprot
H9J3T4
A0A0L7KQC7
A0A2H1WDY9
A0A2A4JFS5
S4NTC9
A0A212F040
+ More
A0A194QE26 A0A0D2X5D9 A0A0D2WX36 A0A2P6SGN3 A0A0D2WUF4 A0A087YQB2 A0A3P9Q2V7 A0A2S2R5K6 A0A3B3YP01 A0A3B5R9Q6 A0A3B5QS14 L1I8L9 A0A315W1G3 A0A0D2UTD6 A0A2P6SHC1 A0A146ZXB2 K7H568 A0A2H8TU71 A0A1W4XAP3 W5NMG6 A0A226EY20 A0A224XH39 A0A1P8C3Y8 A0A0L7LNZ5 A0A1Y1N1E5 A0A3Q3IRY2 A0A1Y1MRB8 A0A2R7WRI7 A0A369RQ21 A0A260YS12 A0A261CFQ0 T1J3Q9 A0A2P4V7H9 E9HEP2 Q18902 J9KX32 A0A2J7R0N6 A0A1B0GPF1 B7P5V9 T1JUP9 T1PHM6 A0A1I8MXN6 A0A1Y1LR74 B3MDZ0 A0A1Y1L0K3 A0A2G5V3R1 A0A087TMU5 A0A0P5SQ96 E2A1E1 A8WTR1 A0A158P3E5 A0A195BID6 L1JYZ8 A0A1B6E1J9 A0A195CD70 A0A210R2N0 A0A147BJ45 A0A195DP96 A0A2H1WSK0 A0A0P6HIA9 A0A164YBP7 E2A9P6 A0A158N8V7 G5CJV8 A0A0P5XDU2 G3PG16 A0A084WJS7 H9J181 A0A0P6CIU7 A0A0P5GME1 D6WRC5 A0A2P6L505 A0A1D1V8K3 A0A182GWJ0 A0A151WLH3 F4WML6 A0A1Y1KTG4 E0VA95 G1NRD1 A0A195CVK3 E9IIM4 A0A2L2YLK1 A0A067R7C0 A0A0J7L688 A0A1Y1KF47 A0A3S2P6L3 A0A0N5CVF8 A0A3Q3MHL5 E0VC27 A0A2J7Q4U1 A0A1W4VZ34 A0A182UJW5 A0A195FNT9
A0A194QE26 A0A0D2X5D9 A0A0D2WX36 A0A2P6SGN3 A0A0D2WUF4 A0A087YQB2 A0A3P9Q2V7 A0A2S2R5K6 A0A3B3YP01 A0A3B5R9Q6 A0A3B5QS14 L1I8L9 A0A315W1G3 A0A0D2UTD6 A0A2P6SHC1 A0A146ZXB2 K7H568 A0A2H8TU71 A0A1W4XAP3 W5NMG6 A0A226EY20 A0A224XH39 A0A1P8C3Y8 A0A0L7LNZ5 A0A1Y1N1E5 A0A3Q3IRY2 A0A1Y1MRB8 A0A2R7WRI7 A0A369RQ21 A0A260YS12 A0A261CFQ0 T1J3Q9 A0A2P4V7H9 E9HEP2 Q18902 J9KX32 A0A2J7R0N6 A0A1B0GPF1 B7P5V9 T1JUP9 T1PHM6 A0A1I8MXN6 A0A1Y1LR74 B3MDZ0 A0A1Y1L0K3 A0A2G5V3R1 A0A087TMU5 A0A0P5SQ96 E2A1E1 A8WTR1 A0A158P3E5 A0A195BID6 L1JYZ8 A0A1B6E1J9 A0A195CD70 A0A210R2N0 A0A147BJ45 A0A195DP96 A0A2H1WSK0 A0A0P6HIA9 A0A164YBP7 E2A9P6 A0A158N8V7 G5CJV8 A0A0P5XDU2 G3PG16 A0A084WJS7 H9J181 A0A0P6CIU7 A0A0P5GME1 D6WRC5 A0A2P6L505 A0A1D1V8K3 A0A182GWJ0 A0A151WLH3 F4WML6 A0A1Y1KTG4 E0VA95 G1NRD1 A0A195CVK3 E9IIM4 A0A2L2YLK1 A0A067R7C0 A0A0J7L688 A0A1Y1KF47 A0A3S2P6L3 A0A0N5CVF8 A0A3Q3MHL5 E0VC27 A0A2J7Q4U1 A0A1W4VZ34 A0A182UJW5 A0A195FNT9
Pubmed
19121390
26227816
23622113
22118469
26354079
23542700
+ More
23201678 29703783 28004739 30042472 26114425 26394399 21292972 9851916 25315136 17994087 20798317 14624247 21347285 28812685 29652888 21827783 24438588 18362917 19820115 27649274 26483478 21719571 20566863 20838655 21282665 26561354 24845553
23201678 29703783 28004739 30042472 26114425 26394399 21292972 9851916 25315136 17994087 20798317 14624247 21347285 28812685 29652888 21827783 24438588 18362917 19820115 27649274 26483478 21719571 20566863 20838655 21282665 26561354 24845553
EMBL
BABH01028786
JTDY01007346
KOB65276.1
ODYU01007994
SOQ51237.1
NWSH01001571
+ More
PCG70831.1 GAIX01012246 JAA80314.1 AGBW02011173 OWR47103.1 KQ459185 KPJ03200.1 KE346375 KJE97639.1 KJE97645.1 PDCK01000039 PRQ57855.1 KE346371 KJE96300.1 AYCK01004920 GGMS01016031 MBY85234.1 JH993190 EKX32427.1 NHOQ01001253 PWA25543.1 KE346377 KJE98246.1 PRQ58036.1 GCES01015363 JAR70960.1 GFXV01005556 MBW17361.1 AHAT01023842 LNIX01000001 OXA61496.1 GFTR01008534 JAW07892.1 KX424937 AOF79114.1 JTDY01000441 KOB77157.1 GEZM01020144 JAV89367.1 GEZM01023766 JAV88201.1 KK855360 PTY22178.1 NOWV01000313 RDD36795.1 NMWX01000827 OZF76206.1 NIPN01000010 OZG21088.1 JH431830 LFJK02001230 POM33246.1 GL732630 EFX69810.1 BX284602 CCD68208.1 ABLF02007606 ABLF02009963 ABLF02009965 NEVH01008227 PNF34397.1 AJVK01059611 ABJB011066243 DS642947 EEC01981.1 CAEY01000780 KA648271 AFP62900.1 GEZM01052581 JAV74405.1 CH902619 EDV37535.2 GEZM01070752 JAV66301.1 PDUG01000002 PIC46380.1 KK115950 KFM66434.1 GDIP01141922 JAL61792.1 GL435760 EFN72767.1 HE601438 CAP23873.1 ADTU01007718 KQ976467 KYM84132.1 JH992969 EKX53569.1 GEDC01005531 JAS31767.1 KQ977935 KYM98660.1 NEDP02000736 OWF55204.1 GEGO01004595 JAR90809.1 KQ980713 KYN14309.1 ODYU01010707 SOQ55987.1 GDIQ01019218 JAN75519.1 LRGB01000930 KZS15088.1 GL437918 EFN69848.1 ADTU01008823 JN180638 AEK86517.1 GDIP01075128 JAM28587.1 ATLV01024054 KE525348 KFB50471.1 BABH01031438 GDIP01007816 JAM95899.1 GDIQ01238327 JAK13398.1 KQ971351 EFA07490.2 MWRG01001781 PRD33665.1 BDGG01000003 GAU96017.1 JXUM01093527 KQ564062 KXJ72853.1 KQ982959 KYQ48657.1 GL888218 EGI64493.1 GEZM01078441 JAV62706.1 DS235005 EEB10301.1 KQ977279 KYN04189.1 GL763494 EFZ19579.1 IAAA01037148 LAA08330.1 KK852657 KDR19221.1 LBMM01000515 KMQ98161.1 GEZM01088340 JAV58255.1 RSAL01000011 RVE53642.1 UYYF01004282 VDN01367.1 DS235045 EEB10933.1 NEVH01018377 PNF23602.1 KQ981382 KYN42215.1
PCG70831.1 GAIX01012246 JAA80314.1 AGBW02011173 OWR47103.1 KQ459185 KPJ03200.1 KE346375 KJE97639.1 KJE97645.1 PDCK01000039 PRQ57855.1 KE346371 KJE96300.1 AYCK01004920 GGMS01016031 MBY85234.1 JH993190 EKX32427.1 NHOQ01001253 PWA25543.1 KE346377 KJE98246.1 PRQ58036.1 GCES01015363 JAR70960.1 GFXV01005556 MBW17361.1 AHAT01023842 LNIX01000001 OXA61496.1 GFTR01008534 JAW07892.1 KX424937 AOF79114.1 JTDY01000441 KOB77157.1 GEZM01020144 JAV89367.1 GEZM01023766 JAV88201.1 KK855360 PTY22178.1 NOWV01000313 RDD36795.1 NMWX01000827 OZF76206.1 NIPN01000010 OZG21088.1 JH431830 LFJK02001230 POM33246.1 GL732630 EFX69810.1 BX284602 CCD68208.1 ABLF02007606 ABLF02009963 ABLF02009965 NEVH01008227 PNF34397.1 AJVK01059611 ABJB011066243 DS642947 EEC01981.1 CAEY01000780 KA648271 AFP62900.1 GEZM01052581 JAV74405.1 CH902619 EDV37535.2 GEZM01070752 JAV66301.1 PDUG01000002 PIC46380.1 KK115950 KFM66434.1 GDIP01141922 JAL61792.1 GL435760 EFN72767.1 HE601438 CAP23873.1 ADTU01007718 KQ976467 KYM84132.1 JH992969 EKX53569.1 GEDC01005531 JAS31767.1 KQ977935 KYM98660.1 NEDP02000736 OWF55204.1 GEGO01004595 JAR90809.1 KQ980713 KYN14309.1 ODYU01010707 SOQ55987.1 GDIQ01019218 JAN75519.1 LRGB01000930 KZS15088.1 GL437918 EFN69848.1 ADTU01008823 JN180638 AEK86517.1 GDIP01075128 JAM28587.1 ATLV01024054 KE525348 KFB50471.1 BABH01031438 GDIP01007816 JAM95899.1 GDIQ01238327 JAK13398.1 KQ971351 EFA07490.2 MWRG01001781 PRD33665.1 BDGG01000003 GAU96017.1 JXUM01093527 KQ564062 KXJ72853.1 KQ982959 KYQ48657.1 GL888218 EGI64493.1 GEZM01078441 JAV62706.1 DS235005 EEB10301.1 KQ977279 KYN04189.1 GL763494 EFZ19579.1 IAAA01037148 LAA08330.1 KK852657 KDR19221.1 LBMM01000515 KMQ98161.1 GEZM01088340 JAV58255.1 RSAL01000011 RVE53642.1 UYYF01004282 VDN01367.1 DS235045 EEB10933.1 NEVH01018377 PNF23602.1 KQ981382 KYN42215.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053268
UP000008743
+ More
UP000238479 UP000028760 UP000242638 UP000261480 UP000002852 UP000011087 UP000265000 UP000005237 UP000192223 UP000018468 UP000198287 UP000261600 UP000253843 UP000216624 UP000216463 UP000237256 UP000000305 UP000001940 UP000007819 UP000235965 UP000092462 UP000001555 UP000015104 UP000095301 UP000007801 UP000230233 UP000054359 UP000000311 UP000008549 UP000005205 UP000078540 UP000078542 UP000242188 UP000078492 UP000076858 UP000007635 UP000030765 UP000007266 UP000186922 UP000069940 UP000249989 UP000075809 UP000007755 UP000009046 UP000001645 UP000027135 UP000036403 UP000283053 UP000046394 UP000276776 UP000261640 UP000192221 UP000075902 UP000078541
UP000238479 UP000028760 UP000242638 UP000261480 UP000002852 UP000011087 UP000265000 UP000005237 UP000192223 UP000018468 UP000198287 UP000261600 UP000253843 UP000216624 UP000216463 UP000237256 UP000000305 UP000001940 UP000007819 UP000235965 UP000092462 UP000001555 UP000015104 UP000095301 UP000007801 UP000230233 UP000054359 UP000000311 UP000008549 UP000005205 UP000078540 UP000078542 UP000242188 UP000078492 UP000076858 UP000007635 UP000030765 UP000007266 UP000186922 UP000069940 UP000249989 UP000075809 UP000007755 UP000009046 UP000001645 UP000027135 UP000036403 UP000283053 UP000046394 UP000276776 UP000261640 UP000192221 UP000075902 UP000078541
Pfam
Interpro
IPR003591
Leu-rich_rpt_typical-subtyp
+ More
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR009030 Growth_fac_rcpt_cys_sf
IPR006212 Furin_repeat
IPR026906 LRR_5
IPR008266 Tyr_kinase_AS
IPR000372 LRRNT
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR013210 LRR_N_plant-typ
IPR025875 Leu-rich_rpt_4
IPR000483 Cys-rich_flank_reg_C
IPR000276 GPCR_Rhodpsn
IPR017452 GPCR_Rhodpsn_7TM
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR036116 FN3_sf
IPR017441 Protein_kinase_ATP_BS
IPR000157 TIR_dom
IPR035897 Toll_tir_struct_dom_sf
IPR003961 FN3_dom
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR009030 Growth_fac_rcpt_cys_sf
IPR006212 Furin_repeat
IPR026906 LRR_5
IPR008266 Tyr_kinase_AS
IPR000372 LRRNT
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR013210 LRR_N_plant-typ
IPR025875 Leu-rich_rpt_4
IPR000483 Cys-rich_flank_reg_C
IPR000276 GPCR_Rhodpsn
IPR017452 GPCR_Rhodpsn_7TM
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR036116 FN3_sf
IPR017441 Protein_kinase_ATP_BS
IPR000157 TIR_dom
IPR035897 Toll_tir_struct_dom_sf
IPR003961 FN3_dom
SUPFAM
Gene 3D
ProteinModelPortal
H9J3T4
A0A0L7KQC7
A0A2H1WDY9
A0A2A4JFS5
S4NTC9
A0A212F040
+ More
A0A194QE26 A0A0D2X5D9 A0A0D2WX36 A0A2P6SGN3 A0A0D2WUF4 A0A087YQB2 A0A3P9Q2V7 A0A2S2R5K6 A0A3B3YP01 A0A3B5R9Q6 A0A3B5QS14 L1I8L9 A0A315W1G3 A0A0D2UTD6 A0A2P6SHC1 A0A146ZXB2 K7H568 A0A2H8TU71 A0A1W4XAP3 W5NMG6 A0A226EY20 A0A224XH39 A0A1P8C3Y8 A0A0L7LNZ5 A0A1Y1N1E5 A0A3Q3IRY2 A0A1Y1MRB8 A0A2R7WRI7 A0A369RQ21 A0A260YS12 A0A261CFQ0 T1J3Q9 A0A2P4V7H9 E9HEP2 Q18902 J9KX32 A0A2J7R0N6 A0A1B0GPF1 B7P5V9 T1JUP9 T1PHM6 A0A1I8MXN6 A0A1Y1LR74 B3MDZ0 A0A1Y1L0K3 A0A2G5V3R1 A0A087TMU5 A0A0P5SQ96 E2A1E1 A8WTR1 A0A158P3E5 A0A195BID6 L1JYZ8 A0A1B6E1J9 A0A195CD70 A0A210R2N0 A0A147BJ45 A0A195DP96 A0A2H1WSK0 A0A0P6HIA9 A0A164YBP7 E2A9P6 A0A158N8V7 G5CJV8 A0A0P5XDU2 G3PG16 A0A084WJS7 H9J181 A0A0P6CIU7 A0A0P5GME1 D6WRC5 A0A2P6L505 A0A1D1V8K3 A0A182GWJ0 A0A151WLH3 F4WML6 A0A1Y1KTG4 E0VA95 G1NRD1 A0A195CVK3 E9IIM4 A0A2L2YLK1 A0A067R7C0 A0A0J7L688 A0A1Y1KF47 A0A3S2P6L3 A0A0N5CVF8 A0A3Q3MHL5 E0VC27 A0A2J7Q4U1 A0A1W4VZ34 A0A182UJW5 A0A195FNT9
A0A194QE26 A0A0D2X5D9 A0A0D2WX36 A0A2P6SGN3 A0A0D2WUF4 A0A087YQB2 A0A3P9Q2V7 A0A2S2R5K6 A0A3B3YP01 A0A3B5R9Q6 A0A3B5QS14 L1I8L9 A0A315W1G3 A0A0D2UTD6 A0A2P6SHC1 A0A146ZXB2 K7H568 A0A2H8TU71 A0A1W4XAP3 W5NMG6 A0A226EY20 A0A224XH39 A0A1P8C3Y8 A0A0L7LNZ5 A0A1Y1N1E5 A0A3Q3IRY2 A0A1Y1MRB8 A0A2R7WRI7 A0A369RQ21 A0A260YS12 A0A261CFQ0 T1J3Q9 A0A2P4V7H9 E9HEP2 Q18902 J9KX32 A0A2J7R0N6 A0A1B0GPF1 B7P5V9 T1JUP9 T1PHM6 A0A1I8MXN6 A0A1Y1LR74 B3MDZ0 A0A1Y1L0K3 A0A2G5V3R1 A0A087TMU5 A0A0P5SQ96 E2A1E1 A8WTR1 A0A158P3E5 A0A195BID6 L1JYZ8 A0A1B6E1J9 A0A195CD70 A0A210R2N0 A0A147BJ45 A0A195DP96 A0A2H1WSK0 A0A0P6HIA9 A0A164YBP7 E2A9P6 A0A158N8V7 G5CJV8 A0A0P5XDU2 G3PG16 A0A084WJS7 H9J181 A0A0P6CIU7 A0A0P5GME1 D6WRC5 A0A2P6L505 A0A1D1V8K3 A0A182GWJ0 A0A151WLH3 F4WML6 A0A1Y1KTG4 E0VA95 G1NRD1 A0A195CVK3 E9IIM4 A0A2L2YLK1 A0A067R7C0 A0A0J7L688 A0A1Y1KF47 A0A3S2P6L3 A0A0N5CVF8 A0A3Q3MHL5 E0VC27 A0A2J7Q4U1 A0A1W4VZ34 A0A182UJW5 A0A195FNT9
PDB
4KNG
E-value=1.68075e-18,
Score=229
Ontologies
GO
Topology
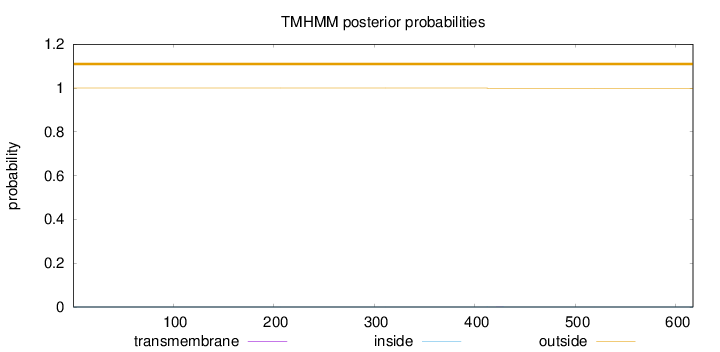
Length:
617
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00424
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00036
outside
1 - 617
Population Genetic Test Statistics
Pi
5.586209
Theta
6.541388
Tajima's D
-0.682427
CLR
0.033489
CSRT
0.205289735513224
Interpretation
Uncertain
