Gene
KWMTBOMO11705
Pre Gene Modal
BGIBMGA004670
Annotation
PREDICTED:_solute_carrier_family_22_member_5-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.97
Sequence
CDS
ATGCAAGAAATTAATGAAGCGTACATCGCATGCGGTTTCGGGTGGTTCCACATCCGGCTACTGGCTATATCGTTTGTGGGCTTCGTGGCGGGGGTTTTGGTCAGCAACAGCACTGCGTACCTGCTCCCCAGTGCCGAATGCGATCTCAACATGTCGTTGGTGCAGAAAGGCTTACTGAATGCTATACCTTACTCAGGAATGCTGCTCTCGACCGTGATAGCCGGATTCCTAACCGACACTTTTGGGAGGAAGACTTTTGTTACTCTCGGATTCGGAGGGATGTTTATATTCATGTTCATTTCGGCTTCAAGCCAAACTTACGAAGTATTGATCACGGCAAAATTCTTCGAGGGTATTCTATTTGCAACATCCTTCAGTGCGGCGGTAACATTGACAACGGAATACTGCCACAATCGAATCAGGGACCAAGTTTTCTTATGCCAGTCGTCTTTCGCCGCTATTGCACAAATTATCATTGCTGCGATGTCATTTGCGATATTAATGAATGATTGGAAATTTAATCTGGGAGGTTTTTTGGAACTGAACACTTGGAATTTTTACCTATATCTGATGTCTTTGTGGTCAGTATCAGCGTGTTTGTTGTACACGATTTTCATTCCCGAAAGTCCAAAATACTTGATAACGCAGAAGAAATTCAACGAAGCGAGGAATATATTAATCAAAATGTATACAATGAATACTGGTAAACCAGAGAACACATTTCCGTACCCAAACTTATGGAAAGATAAGCTGCCACAGCAAGTCGAAGAAGCTTCGAAAGAAAATCTATGCAAAGCTTTTAAACACCAGATTGTCGTCGGACTAAGAAATGTTAAACCAATGTTCAGGAGACCTTTGGTTTTGTATCTGCTAATGATTAGCTTCTCGCAGTTTTTGATAATGGGTGTGTACAACGTATTGAGGTTATGGTATCCTCAATTGTCGACTATTGTCGAGCACTACAGTACGGGAACTAACAGTAATTTGTGTAGCATGTTGGATGCTTACACAGCTGACCTGAAGACCGCTGTGACTAATTCGTCGAGCTCCGACGTCTGTGTGGTGACCCCAAGCGGAGCTGAAACATACATAAACAGCATGATTATTGGCTGCGTCTGTCTCCTACCGTATTTCGTCTCCGGGGTTATCGTCAACCGTGTTGGGAAGAAACCATTGCTGATCGCAGCTGGCGTGATCACTACAGCCATAACTTTAAGCGTTCAATGGGCCAATTCCAAAGCAGCGATCGTATCGCTTTTTGCTATCGATACAGCTGTGGGTCAGGTTTTGTTGAGCTTGAATCAGGCCTTGACAGTTGAGTTGTTTCCGACAACTACTAGAACGTTGGCTATTTCTATGATAATGGTGGCTGGGAGGATCGGTACCTTAACAGCGAATGTGGCGTTCCCGGTTCTCCTTGATATGGGCTGCGGAGTACCATTTTTCACGTTGACCGGAATGGTCATGTGTGTAACTGCGATATCTTTGTTCCTGCCTTCCGCGAAGAAGTAA
Protein
MQEINEAYIACGFGWFHIRLLAISFVGFVAGVLVSNSTAYLLPSAECDLNMSLVQKGLLNAIPYSGMLLSTVIAGFLTDTFGRKTFVTLGFGGMFIFMFISASSQTYEVLITAKFFEGILFATSFSAAVTLTTEYCHNRIRDQVFLCQSSFAAIAQIIIAAMSFAILMNDWKFNLGGFLELNTWNFYLYLMSLWSVSACLLYTIFIPESPKYLITQKKFNEARNILIKMYTMNTGKPENTFPYPNLWKDKLPQQVEEASKENLCKAFKHQIVVGLRNVKPMFRRPLVLYLLMISFSQFLIMGVYNVLRLWYPQLSTIVEHYSTGTNSNLCSMLDAYTADLKTAVTNSSSSDVCVVTPSGAETYINSMIIGCVCLLPYFVSGVIVNRVGKKPLLIAAGVITTAITLSVQWANSKAAIVSLFAIDTAVGQVLLSLNQALTVELFPTTTRTLAISMIMVAGRIGTLTANVAFPVLLDMGCGVPFFTLTGMVMCVTAISLFLPSAKK
Summary
Uniprot
A0A194QCK7
A0A2A4JJY9
A0A194QYS5
A0A2H1VK18
A0A194QIC4
A0A194QY24
+ More
A0A2H1VJ05 A0A194QZP1 A0A2A4JL57 A0A194QCD1 A0A194R5V0 J3JVU6 A0A1I8PCF2 A0A1I8PT34 A0A1I8MPQ8 A0A1I8PT04 D6W8I7 A0A0L0BZW9 A0A1I8NF35 A0A1I8MTP1 A0A139WMI2 A0A0A1XHP3 A0A1I8N9Q0 U4UFR8 B4LZI9 N6UKT9 A0A1I8PX70 A0A067RW60 A0A034WRA7 A0A0K8UTD6 A0A1W4VZN5 A0A1I8NY17 B4KD04 A0A1L8DV70 W8C4Z8 A0A0L0C6H5 A0A2H1V9E4 K7IVE4 A0A139WMH8 A0A0M4F861 A0A139WMN4 A0A3B0KYE1 A0A1I8M6J6 B4MD31 T1PAI7 A0A088AE26 A0A0N0BCS7 A0A2A4JM14 A0A2A3EAK9 B5DL72 A0A2J7RAL7 A0A2J7RAM3 A0A1I8M3K1 B4NK00 N6TIC7 A0A0K8WKA0 U4UDT0 A0A1A9ZAD2 A0A0K8V634 A0A232ETW6 K7IYZ5 A0A310SUH1 B4HIQ9 B3P1K9 B0X4T6 A0A1Q3FPY0 A0A1W4WUS5 A0A1I8PD47 A0A3L8DU11 B4PLC5 A0A034VN09 A0A3B0KC88 B3LWI0 E2BCE5 A0A0A1XIE2 K7IVE2 A0A194Q2G9 B3P9C6 A0A026W3L9 Q9V3J5 D0Z737 A0A232ETP1 A0A0A1XT62 E2ADZ1 B4JKV4 B6IDU4 B4NK01 F4X8U8 B4GTT2 B4PXJ4 B4NK02 A0A0P6IV24 Q293R1 B4GLR6 W8BP25 A0A139WMM0 A0A158NBZ9 Q9VGX6
A0A2H1VJ05 A0A194QZP1 A0A2A4JL57 A0A194QCD1 A0A194R5V0 J3JVU6 A0A1I8PCF2 A0A1I8PT34 A0A1I8MPQ8 A0A1I8PT04 D6W8I7 A0A0L0BZW9 A0A1I8NF35 A0A1I8MTP1 A0A139WMI2 A0A0A1XHP3 A0A1I8N9Q0 U4UFR8 B4LZI9 N6UKT9 A0A1I8PX70 A0A067RW60 A0A034WRA7 A0A0K8UTD6 A0A1W4VZN5 A0A1I8NY17 B4KD04 A0A1L8DV70 W8C4Z8 A0A0L0C6H5 A0A2H1V9E4 K7IVE4 A0A139WMH8 A0A0M4F861 A0A139WMN4 A0A3B0KYE1 A0A1I8M6J6 B4MD31 T1PAI7 A0A088AE26 A0A0N0BCS7 A0A2A4JM14 A0A2A3EAK9 B5DL72 A0A2J7RAL7 A0A2J7RAM3 A0A1I8M3K1 B4NK00 N6TIC7 A0A0K8WKA0 U4UDT0 A0A1A9ZAD2 A0A0K8V634 A0A232ETW6 K7IYZ5 A0A310SUH1 B4HIQ9 B3P1K9 B0X4T6 A0A1Q3FPY0 A0A1W4WUS5 A0A1I8PD47 A0A3L8DU11 B4PLC5 A0A034VN09 A0A3B0KC88 B3LWI0 E2BCE5 A0A0A1XIE2 K7IVE2 A0A194Q2G9 B3P9C6 A0A026W3L9 Q9V3J5 D0Z737 A0A232ETP1 A0A0A1XT62 E2ADZ1 B4JKV4 B6IDU4 B4NK01 F4X8U8 B4GTT2 B4PXJ4 B4NK02 A0A0P6IV24 Q293R1 B4GLR6 W8BP25 A0A139WMM0 A0A158NBZ9 Q9VGX6
Pubmed
EMBL
KQ459185
KPJ03202.1
NWSH01001203
PCG72139.1
KQ460949
KPJ10444.1
+ More
ODYU01002822 SOQ40782.1 KPJ03201.1 KPJ10443.1 SOQ40781.1 KPJ10445.1 PCG72140.1 KPJ03203.1 KQ460685 KPJ12889.1 BT127364 AEE62326.1 KQ971312 EEZ98361.1 JRES01001097 KNC25563.1 KYB29086.1 GBXI01003857 JAD10435.1 KB632277 ERL91213.1 CH940650 EDW67128.1 APGK01019806 APGK01019807 KB740128 ENN81286.1 KK852427 KDR24109.1 GAKP01002262 JAC56690.1 GDHF01022357 JAI29957.1 CH933806 EDW13774.1 GFDF01003899 JAV10185.1 GAMC01009291 JAB97264.1 JRES01000940 KNC27034.1 ODYU01001382 SOQ37473.1 AAZX01005017 KYB29084.1 CP012528 ALC48157.1 KYB29085.1 OUUW01000018 SPP89118.1 CH940660 EDW58103.1 KA645786 AFP60415.1 KQ435900 KOX69091.1 NWSH01001073 PCG72758.1 KZ288315 PBC28332.1 CH379063 EDY72271.1 NEVH01006567 PNF37879.1 PNF37878.1 CH964272 EDW84002.1 APGK01025268 APGK01025269 APGK01025270 KB740583 ENN80174.1 GDHF01000738 JAI51576.1 ERL91212.1 GDHF01018254 JAI34060.1 NNAY01002245 OXU21736.1 KQ760133 OAD61788.1 CH480815 EDW42706.1 CH954181 EDV49608.1 DS232352 EDS40551.1 GFDL01005483 JAV29562.1 QOIP01000004 RLU23874.1 CM000160 EDW96832.1 GAKP01015787 JAC43165.1 OUUW01000008 SPP83799.1 CH902617 EDV41574.1 GL447257 EFN86681.1 GBXI01003143 JAD11149.1 KQ459579 KPI99533.1 CH954183 EDV45422.2 KK107453 EZA50652.1 AE014298 AL109630 AAF45574.3 CAB65877.1 BT100284 ACY79579.1 NNAY01002248 OXU21728.1 GBXI01000534 JAD13758.1 GL438827 EFN68224.1 CH916370 EDW00207.1 BT050534 ACJ13241.1 EDW84003.2 GL888932 EGI57367.1 CH479190 EDW25952.1 CM000162 EDX00847.1 EDW84004.1 GDUN01000918 JAN95001.1 CM000070 EAL29153.2 CH479185 EDW38490.1 GAMC01005923 JAC00633.1 KYB29087.1 ADTU01010989 ADTU01010990 ADTU01010991 AE014297 BT044142 AAF54546.3 ACH92207.1
ODYU01002822 SOQ40782.1 KPJ03201.1 KPJ10443.1 SOQ40781.1 KPJ10445.1 PCG72140.1 KPJ03203.1 KQ460685 KPJ12889.1 BT127364 AEE62326.1 KQ971312 EEZ98361.1 JRES01001097 KNC25563.1 KYB29086.1 GBXI01003857 JAD10435.1 KB632277 ERL91213.1 CH940650 EDW67128.1 APGK01019806 APGK01019807 KB740128 ENN81286.1 KK852427 KDR24109.1 GAKP01002262 JAC56690.1 GDHF01022357 JAI29957.1 CH933806 EDW13774.1 GFDF01003899 JAV10185.1 GAMC01009291 JAB97264.1 JRES01000940 KNC27034.1 ODYU01001382 SOQ37473.1 AAZX01005017 KYB29084.1 CP012528 ALC48157.1 KYB29085.1 OUUW01000018 SPP89118.1 CH940660 EDW58103.1 KA645786 AFP60415.1 KQ435900 KOX69091.1 NWSH01001073 PCG72758.1 KZ288315 PBC28332.1 CH379063 EDY72271.1 NEVH01006567 PNF37879.1 PNF37878.1 CH964272 EDW84002.1 APGK01025268 APGK01025269 APGK01025270 KB740583 ENN80174.1 GDHF01000738 JAI51576.1 ERL91212.1 GDHF01018254 JAI34060.1 NNAY01002245 OXU21736.1 KQ760133 OAD61788.1 CH480815 EDW42706.1 CH954181 EDV49608.1 DS232352 EDS40551.1 GFDL01005483 JAV29562.1 QOIP01000004 RLU23874.1 CM000160 EDW96832.1 GAKP01015787 JAC43165.1 OUUW01000008 SPP83799.1 CH902617 EDV41574.1 GL447257 EFN86681.1 GBXI01003143 JAD11149.1 KQ459579 KPI99533.1 CH954183 EDV45422.2 KK107453 EZA50652.1 AE014298 AL109630 AAF45574.3 CAB65877.1 BT100284 ACY79579.1 NNAY01002248 OXU21728.1 GBXI01000534 JAD13758.1 GL438827 EFN68224.1 CH916370 EDW00207.1 BT050534 ACJ13241.1 EDW84003.2 GL888932 EGI57367.1 CH479190 EDW25952.1 CM000162 EDX00847.1 EDW84004.1 GDUN01000918 JAN95001.1 CM000070 EAL29153.2 CH479185 EDW38490.1 GAMC01005923 JAC00633.1 KYB29087.1 ADTU01010989 ADTU01010990 ADTU01010991 AE014297 BT044142 AAF54546.3 ACH92207.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000095300
UP000095301
UP000007266
+ More
UP000037069 UP000030742 UP000008792 UP000019118 UP000027135 UP000192221 UP000009192 UP000002358 UP000092553 UP000268350 UP000005203 UP000053105 UP000242457 UP000001819 UP000235965 UP000007798 UP000092445 UP000215335 UP000001292 UP000008711 UP000002320 UP000192223 UP000279307 UP000002282 UP000007801 UP000008237 UP000053097 UP000000803 UP000000311 UP000001070 UP000007755 UP000008744 UP000005205
UP000037069 UP000030742 UP000008792 UP000019118 UP000027135 UP000192221 UP000009192 UP000002358 UP000092553 UP000268350 UP000005203 UP000053105 UP000242457 UP000001819 UP000235965 UP000007798 UP000092445 UP000215335 UP000001292 UP000008711 UP000002320 UP000192223 UP000279307 UP000002282 UP000007801 UP000008237 UP000053097 UP000000803 UP000000311 UP000001070 UP000007755 UP000008744 UP000005205
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A194QCK7
A0A2A4JJY9
A0A194QYS5
A0A2H1VK18
A0A194QIC4
A0A194QY24
+ More
A0A2H1VJ05 A0A194QZP1 A0A2A4JL57 A0A194QCD1 A0A194R5V0 J3JVU6 A0A1I8PCF2 A0A1I8PT34 A0A1I8MPQ8 A0A1I8PT04 D6W8I7 A0A0L0BZW9 A0A1I8NF35 A0A1I8MTP1 A0A139WMI2 A0A0A1XHP3 A0A1I8N9Q0 U4UFR8 B4LZI9 N6UKT9 A0A1I8PX70 A0A067RW60 A0A034WRA7 A0A0K8UTD6 A0A1W4VZN5 A0A1I8NY17 B4KD04 A0A1L8DV70 W8C4Z8 A0A0L0C6H5 A0A2H1V9E4 K7IVE4 A0A139WMH8 A0A0M4F861 A0A139WMN4 A0A3B0KYE1 A0A1I8M6J6 B4MD31 T1PAI7 A0A088AE26 A0A0N0BCS7 A0A2A4JM14 A0A2A3EAK9 B5DL72 A0A2J7RAL7 A0A2J7RAM3 A0A1I8M3K1 B4NK00 N6TIC7 A0A0K8WKA0 U4UDT0 A0A1A9ZAD2 A0A0K8V634 A0A232ETW6 K7IYZ5 A0A310SUH1 B4HIQ9 B3P1K9 B0X4T6 A0A1Q3FPY0 A0A1W4WUS5 A0A1I8PD47 A0A3L8DU11 B4PLC5 A0A034VN09 A0A3B0KC88 B3LWI0 E2BCE5 A0A0A1XIE2 K7IVE2 A0A194Q2G9 B3P9C6 A0A026W3L9 Q9V3J5 D0Z737 A0A232ETP1 A0A0A1XT62 E2ADZ1 B4JKV4 B6IDU4 B4NK01 F4X8U8 B4GTT2 B4PXJ4 B4NK02 A0A0P6IV24 Q293R1 B4GLR6 W8BP25 A0A139WMM0 A0A158NBZ9 Q9VGX6
A0A2H1VJ05 A0A194QZP1 A0A2A4JL57 A0A194QCD1 A0A194R5V0 J3JVU6 A0A1I8PCF2 A0A1I8PT34 A0A1I8MPQ8 A0A1I8PT04 D6W8I7 A0A0L0BZW9 A0A1I8NF35 A0A1I8MTP1 A0A139WMI2 A0A0A1XHP3 A0A1I8N9Q0 U4UFR8 B4LZI9 N6UKT9 A0A1I8PX70 A0A067RW60 A0A034WRA7 A0A0K8UTD6 A0A1W4VZN5 A0A1I8NY17 B4KD04 A0A1L8DV70 W8C4Z8 A0A0L0C6H5 A0A2H1V9E4 K7IVE4 A0A139WMH8 A0A0M4F861 A0A139WMN4 A0A3B0KYE1 A0A1I8M6J6 B4MD31 T1PAI7 A0A088AE26 A0A0N0BCS7 A0A2A4JM14 A0A2A3EAK9 B5DL72 A0A2J7RAL7 A0A2J7RAM3 A0A1I8M3K1 B4NK00 N6TIC7 A0A0K8WKA0 U4UDT0 A0A1A9ZAD2 A0A0K8V634 A0A232ETW6 K7IYZ5 A0A310SUH1 B4HIQ9 B3P1K9 B0X4T6 A0A1Q3FPY0 A0A1W4WUS5 A0A1I8PD47 A0A3L8DU11 B4PLC5 A0A034VN09 A0A3B0KC88 B3LWI0 E2BCE5 A0A0A1XIE2 K7IVE2 A0A194Q2G9 B3P9C6 A0A026W3L9 Q9V3J5 D0Z737 A0A232ETP1 A0A0A1XT62 E2ADZ1 B4JKV4 B6IDU4 B4NK01 F4X8U8 B4GTT2 B4PXJ4 B4NK02 A0A0P6IV24 Q293R1 B4GLR6 W8BP25 A0A139WMM0 A0A158NBZ9 Q9VGX6
PDB
4LDS
E-value=0.0677676,
Score=85
Ontologies
GO
Topology
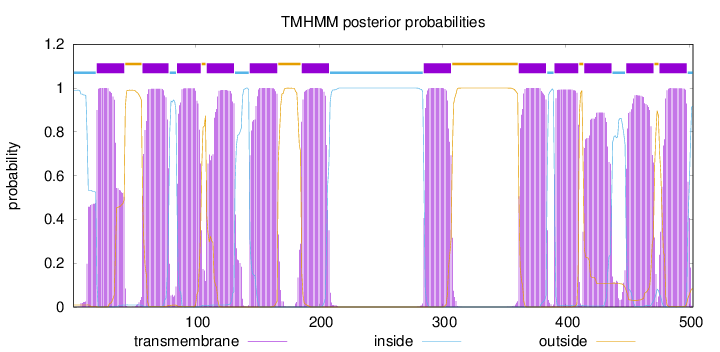
Length:
503
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
255.89639
Exp number, first 60 AAs:
25.83415
Total prob of N-in:
0.99033
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 56
TMhelix
57 - 78
inside
79 - 84
TMhelix
85 - 104
outside
105 - 108
TMhelix
109 - 131
inside
132 - 143
TMhelix
144 - 166
outside
167 - 185
TMhelix
186 - 208
inside
209 - 284
TMhelix
285 - 307
outside
308 - 361
TMhelix
362 - 384
inside
385 - 390
TMhelix
391 - 410
outside
411 - 414
TMhelix
415 - 437
inside
438 - 448
TMhelix
449 - 471
outside
472 - 475
TMhelix
476 - 498
inside
499 - 503
Population Genetic Test Statistics
Pi
32.026999
Theta
30.49442
Tajima's D
0.168877
CLR
1.037335
CSRT
0.416129193540323
Interpretation
Uncertain
