Gene
KWMTBOMO11700
Pre Gene Modal
BGIBMGA003965
Annotation
PREDICTED:_hemicentin-2-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.042 PlasmaMembrane Reliability : 1.579
Sequence
CDS
ATGCACTTAACGGAGTGCGATTTAACGTTCTCTTTCAATTGGTTCCAGATGAAATGGAAACGTACAGGCGGAATGGGAATACAGCTAGCTGCAGCAGTGTTCTGGATGCTGCCTTTGATCACGTCTTCAACGGGAGATATAAGGCACCACGGAGCGTCGAGGGCAAGGGGTTGGCGTGCAGGGACCCAGCAGACTGTTGAAACGGCCGCCAATAACTCGGTCAGCAACATCACTGCACAACTAGGCGGAACCGCCTACCTCCACTGTCTAGTTGGGCATCTCAGCGAGAGAGGGCAAGTCTCCTGGATCAGGAAACGTGACTGGCACATTTTGAGCTCCGGGAAATTTACTTACACGAACGACGAGAGATTTCAGGTATTACACGGCGAAGGTTCTGAAGATTGGACTCTGCAGATTAAGTTTGTTCAAAAGAGAGACAACGGTACTTACGAGTGTCAGGTGTCAACAAGAACTGGGATAGTGTCGCATTACGTACGTCTCCAGATAGTGGTTCCGGAGGCCTTCATTCTTGGCTCCGGAGAACACCACGTCGATCTGGGCAGCGTCATCCACCTCATATGTATCGTGGAGAAGAGTCCAACACCCCCGCAATACGTCTTCTGGTACCACAATGACAGGATGATAAACTATGATGCTACAAGAGGAGGGATCACAGTGGAGACCGAGCCCGGCGCCCGTACCCAATCCAGACTCACAGTCAGAGGCGCAACACTCAAAGATTCCGGGAATTACACATGCAGCGCTAGTAACACTGAGCCTGCTTCTATACACGTTTTCGTCTCGGAAGGAAGTAACAAAATGGCGGCGATACTTCGGCGCAACACGAGCGCGATCCACGGGATCACCACTAGCGCCACCTGTCTGCTCCTTCTGGGCACAGCGCTCATGCACTATGGTTTGCGATGA
Protein
MHLTECDLTFSFNWFQMKWKRTGGMGIQLAAAVFWMLPLITSSTGDIRHHGASRARGWRAGTQQTVETAANNSVSNITAQLGGTAYLHCLVGHLSERGQVSWIRKRDWHILSSGKFTYTNDERFQVLHGEGSEDWTLQIKFVQKRDNGTYECQVSTRTGIVSHYVRLQIVVPEAFILGSGEHHVDLGSVIHLICIVEKSPTPPQYVFWYHNDRMINYDATRGGITVETEPGARTQSRLTVRGATLKDSGNYTCSASNTEPASIHVFVSEGSNKMAAILRRNTSAIHGITTSATCLLLLGTALMHYGLR
Summary
Uniprot
A0A3S2NFI6
A0A194QE31
A0A212FNL4
A0A2A4JJL4
A0A0L7L3H1
A0A1B6M5C2
+ More
A0A0C9QGT5 A0A158NDS5 K7J5J1 A0A3L8E1U2 A0A067QY67 A0A088ATH3 A0A0L7R892 F4W811 A0A195EY83 A0A195BQJ6 A0A146KVD1 A0A0A9WHQ0 A0A154PAT5 A0A1Y1KG40 A0A0N0U7W7 A0A2W1C1H9 A0A212FHY4 B4II15 A0A2Y9D2C3 B4NYX1 B3N3H9 A0A0J9R642 A0A0J9TU05 A0A0B4KED6 A0A0P9C121 A0A0Q9X7A2 B4MDZ1 A0A1W4W4S0 A0A1W4VSQ1 A0A0Q9W018 A0A1I8MQG3 A0A1I8QCQ8 A0A0R1DN06 A0A0Q5WCF0 A0A0M4EHL1 A0A0R1DNB1 A0A0P8ZS53 B5DZ10 A0A0J9R7J7 A1Z6H9 A0A0J9R6D3 E3CTN6 A0A0Q9W020 A0A1W4W5B7 A0A1W4VSJ7 A0A0Q9X729 A0A0Q9WCB0 B4KSB9 A0A1B0AC53 A0A1I8QCP5 A0A3B0J6W5 A0A1I8QCM5 A0A1B0G748 A0A1I8QCP4 A0A1I8MQG4 A0A1I8QCU7 A0A1A9V5D0 A0A1B0CLM6 A0A1B6L536 A0A1B6CMM9 A0A194PM23
A0A0C9QGT5 A0A158NDS5 K7J5J1 A0A3L8E1U2 A0A067QY67 A0A088ATH3 A0A0L7R892 F4W811 A0A195EY83 A0A195BQJ6 A0A146KVD1 A0A0A9WHQ0 A0A154PAT5 A0A1Y1KG40 A0A0N0U7W7 A0A2W1C1H9 A0A212FHY4 B4II15 A0A2Y9D2C3 B4NYX1 B3N3H9 A0A0J9R642 A0A0J9TU05 A0A0B4KED6 A0A0P9C121 A0A0Q9X7A2 B4MDZ1 A0A1W4W4S0 A0A1W4VSQ1 A0A0Q9W018 A0A1I8MQG3 A0A1I8QCQ8 A0A0R1DN06 A0A0Q5WCF0 A0A0M4EHL1 A0A0R1DNB1 A0A0P8ZS53 B5DZ10 A0A0J9R7J7 A1Z6H9 A0A0J9R6D3 E3CTN6 A0A0Q9W020 A0A1W4W5B7 A0A1W4VSJ7 A0A0Q9X729 A0A0Q9WCB0 B4KSB9 A0A1B0AC53 A0A1I8QCP5 A0A3B0J6W5 A0A1I8QCM5 A0A1B0G748 A0A1I8QCP4 A0A1I8MQG4 A0A1I8QCU7 A0A1A9V5D0 A0A1B0CLM6 A0A1B6L536 A0A1B6CMM9 A0A194PM23
Pubmed
EMBL
RSAL01000136
RVE46214.1
KQ459185
KPJ03205.1
AGBW02005277
OWR55299.1
+ More
NWSH01001235 PCG72019.1 JTDY01003273 KOB69829.1 GEBQ01008874 GEBQ01007244 JAT31103.1 JAT32733.1 GBYB01013793 JAG83560.1 ADTU01012914 ADTU01012915 ADTU01012916 ADTU01012917 ADTU01012918 ADTU01012919 ADTU01012920 ADTU01012921 ADTU01012922 ADTU01012923 QOIP01000002 RLU26159.1 KK852824 KDR15426.1 KQ414637 KOC66976.1 GL887888 EGI69681.1 KQ981906 KYN33245.1 KQ976424 KYM88408.1 GDHC01018780 JAP99848.1 GBHO01035642 GBHO01031866 GDHC01008910 JAG07962.1 JAG11738.1 JAQ09719.1 KQ434849 KZC08494.1 GEZM01086326 GEZM01086325 GEZM01086323 JAV59548.1 KQ435694 KOX80921.1 KZ149915 PZC77963.1 AGBW02008458 OWR53326.1 CH480841 EDW49541.1 AAAB01008960 CM000157 EDW89822.2 CH954177 EDV59861.2 CM002911 KMY91662.1 KMY91665.1 AE013599 AGB93251.1 CH902619 KPU77310.1 CH933808 KRG04108.1 CH940662 EDW58756.1 KRF78457.1 KRJ98731.1 KQS71043.1 CP012524 ALC40430.1 KRJ98732.1 KPU77311.1 CM000071 EDY69227.3 KMY91664.1 BT029948 AAF57304.3 ABM92822.1 KMY91663.1 BT125702 ADO33193.1 KRF78458.1 KRG04107.1 KRF78459.1 EDW08401.2 OUUW01000001 SPP75823.1 CCAG010003785 AJWK01017457 GEBQ01021373 JAT18604.1 GEDC01022653 JAS14645.1 KQ459598 KPI94486.1
NWSH01001235 PCG72019.1 JTDY01003273 KOB69829.1 GEBQ01008874 GEBQ01007244 JAT31103.1 JAT32733.1 GBYB01013793 JAG83560.1 ADTU01012914 ADTU01012915 ADTU01012916 ADTU01012917 ADTU01012918 ADTU01012919 ADTU01012920 ADTU01012921 ADTU01012922 ADTU01012923 QOIP01000002 RLU26159.1 KK852824 KDR15426.1 KQ414637 KOC66976.1 GL887888 EGI69681.1 KQ981906 KYN33245.1 KQ976424 KYM88408.1 GDHC01018780 JAP99848.1 GBHO01035642 GBHO01031866 GDHC01008910 JAG07962.1 JAG11738.1 JAQ09719.1 KQ434849 KZC08494.1 GEZM01086326 GEZM01086325 GEZM01086323 JAV59548.1 KQ435694 KOX80921.1 KZ149915 PZC77963.1 AGBW02008458 OWR53326.1 CH480841 EDW49541.1 AAAB01008960 CM000157 EDW89822.2 CH954177 EDV59861.2 CM002911 KMY91662.1 KMY91665.1 AE013599 AGB93251.1 CH902619 KPU77310.1 CH933808 KRG04108.1 CH940662 EDW58756.1 KRF78457.1 KRJ98731.1 KQS71043.1 CP012524 ALC40430.1 KRJ98732.1 KPU77311.1 CM000071 EDY69227.3 KMY91664.1 BT029948 AAF57304.3 ABM92822.1 KMY91663.1 BT125702 ADO33193.1 KRF78458.1 KRG04107.1 KRF78459.1 EDW08401.2 OUUW01000001 SPP75823.1 CCAG010003785 AJWK01017457 GEBQ01021373 JAT18604.1 GEDC01022653 JAS14645.1 KQ459598 KPI94486.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000218220
UP000037510
UP000005205
+ More
UP000002358 UP000279307 UP000027135 UP000005203 UP000053825 UP000007755 UP000078541 UP000078540 UP000076502 UP000053105 UP000001292 UP000002282 UP000008711 UP000000803 UP000007801 UP000009192 UP000008792 UP000192221 UP000095301 UP000095300 UP000092553 UP000001819 UP000092445 UP000268350 UP000092444 UP000078200 UP000092461
UP000002358 UP000279307 UP000027135 UP000005203 UP000053825 UP000007755 UP000078541 UP000078540 UP000076502 UP000053105 UP000001292 UP000002282 UP000008711 UP000000803 UP000007801 UP000009192 UP000008792 UP000192221 UP000095301 UP000095300 UP000092553 UP000001819 UP000092445 UP000268350 UP000092444 UP000078200 UP000092461
PRIDE
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A3S2NFI6
A0A194QE31
A0A212FNL4
A0A2A4JJL4
A0A0L7L3H1
A0A1B6M5C2
+ More
A0A0C9QGT5 A0A158NDS5 K7J5J1 A0A3L8E1U2 A0A067QY67 A0A088ATH3 A0A0L7R892 F4W811 A0A195EY83 A0A195BQJ6 A0A146KVD1 A0A0A9WHQ0 A0A154PAT5 A0A1Y1KG40 A0A0N0U7W7 A0A2W1C1H9 A0A212FHY4 B4II15 A0A2Y9D2C3 B4NYX1 B3N3H9 A0A0J9R642 A0A0J9TU05 A0A0B4KED6 A0A0P9C121 A0A0Q9X7A2 B4MDZ1 A0A1W4W4S0 A0A1W4VSQ1 A0A0Q9W018 A0A1I8MQG3 A0A1I8QCQ8 A0A0R1DN06 A0A0Q5WCF0 A0A0M4EHL1 A0A0R1DNB1 A0A0P8ZS53 B5DZ10 A0A0J9R7J7 A1Z6H9 A0A0J9R6D3 E3CTN6 A0A0Q9W020 A0A1W4W5B7 A0A1W4VSJ7 A0A0Q9X729 A0A0Q9WCB0 B4KSB9 A0A1B0AC53 A0A1I8QCP5 A0A3B0J6W5 A0A1I8QCM5 A0A1B0G748 A0A1I8QCP4 A0A1I8MQG4 A0A1I8QCU7 A0A1A9V5D0 A0A1B0CLM6 A0A1B6L536 A0A1B6CMM9 A0A194PM23
A0A0C9QGT5 A0A158NDS5 K7J5J1 A0A3L8E1U2 A0A067QY67 A0A088ATH3 A0A0L7R892 F4W811 A0A195EY83 A0A195BQJ6 A0A146KVD1 A0A0A9WHQ0 A0A154PAT5 A0A1Y1KG40 A0A0N0U7W7 A0A2W1C1H9 A0A212FHY4 B4II15 A0A2Y9D2C3 B4NYX1 B3N3H9 A0A0J9R642 A0A0J9TU05 A0A0B4KED6 A0A0P9C121 A0A0Q9X7A2 B4MDZ1 A0A1W4W4S0 A0A1W4VSQ1 A0A0Q9W018 A0A1I8MQG3 A0A1I8QCQ8 A0A0R1DN06 A0A0Q5WCF0 A0A0M4EHL1 A0A0R1DNB1 A0A0P8ZS53 B5DZ10 A0A0J9R7J7 A1Z6H9 A0A0J9R6D3 E3CTN6 A0A0Q9W020 A0A1W4W5B7 A0A1W4VSJ7 A0A0Q9X729 A0A0Q9WCB0 B4KSB9 A0A1B0AC53 A0A1I8QCP5 A0A3B0J6W5 A0A1I8QCM5 A0A1B0G748 A0A1I8QCP4 A0A1I8MQG4 A0A1I8QCU7 A0A1A9V5D0 A0A1B0CLM6 A0A1B6L536 A0A1B6CMM9 A0A194PM23
PDB
6EG0
E-value=2.11639e-46,
Score=467
Ontologies
GO
PANTHER
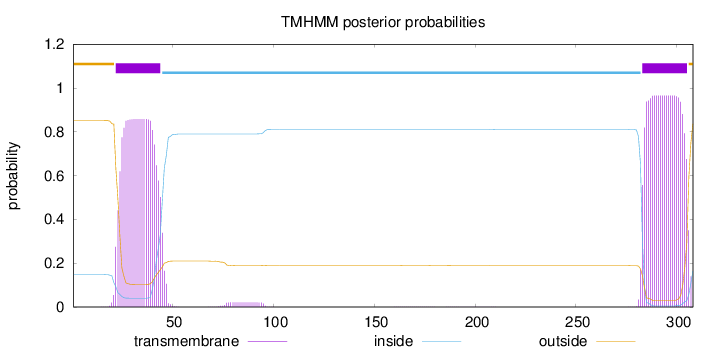
Topology
Length:
308
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.21269
Exp number, first 60 AAs:
18.06503
Total prob of N-in:
0.14810
POSSIBLE N-term signal
sequence
outside
1 - 21
TMhelix
22 - 44
inside
45 - 282
TMhelix
283 - 305
outside
306 - 308
Population Genetic Test Statistics
Pi
25.309885
Theta
22.930058
Tajima's D
0.32358
CLR
1.463165
CSRT
0.463926803659817
Interpretation
Uncertain
