Pre Gene Modal
BGIBMGA004142
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_B_member_8?_mitochondrial_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.059
Sequence
CDS
ATGCGGACGGTTCGAGCCTTCGCCGCCGAAGACAGCGAGGCGCAGATGTTTGCCAAGGAGTGTGACGAGGCCGCCCAGCTCAGCATGGAGCTGGGACTCGGCATCGGATTGTTTCAGGCCGGGACTAACCTGTTCTTGAATGGAATGGTGCTCGGCACTATGTTCCTCGGCGGTCATTTGATGTCGACCGGGCAGATGTCGGCGGGAGACGTGATGGCGTTCCTGGTGAACGCGCAGACTGTGCAGCGCTCGGTGGCGCAGCTGTCTCTGCTGTTCGGCTCCGTCGTTAAGGGGATCAGCGCCGGCGGTCGGGTGTTCGAGTACATCAATAAGAAACCCGCAATGGACACGTCCGGTACTAAGTATATAAAGTACCACGAGTTTCACGGCGACATCGAGTTTAAGAACGTCACATTCGCCTATCCCACAAGACCGGAGGCTGTGATATTGAAGGATTTCAATCTTAAAATACCTGCTGGCAAGACGGTCGCCATAGTCGGCACATCTGGCAACGGGAAGTCTACTATCGCTGCTCTGTTGGAAAGGTTCTACGACGTGAACGAAGGCCAAGTGTCGATAGATGGCGTTGTCTTGCGCGAGTTCGACGGCAAGTGGCTCCGCGGGAGGGCGCTAGGTCTGATCGGCCAGGAGCCGGTCCTGTTCGCCACCACGGTCCGGGAGAACATCCGGTACGGGAGACCAGAGGCCACTGATGCTGAGGTGATGCAGGCGGCTACGATAGCCAATGCTCATGAGTTCATACAGTCGTTCCCGGAGGGGTACGATACTCTGGTCGGCGAGAGGGGGATGGCTCTGAGCGGGGGACAGAAGCAGCGAGTCGCCATCGCCAGAGCCGTGCTGAAGAATCCACCGGTTATAATACTGGACGAGGCCACCAGCGCTCTTGATTCCGCGAGCGAGAAGGTCGTACAGAAGGCGCTGGAGACGGCGGCCAAGGGTCGGACCGTGCTGGTGATAGCCCACAGGCTATCCACGGTCATCAATGCCGATCTGATCGTCGTATTAAACAAGGGGAAGATTGTTGAGATGGGAAATCATGAACAATTGAAGAAATTGAAGGGTTTTTATTGGAATCTTATAAAGCAACAGGATCAAGATCTAGAGCAATCGAGATGA
Protein
MRTVRAFAAEDSEAQMFAKECDEAAQLSMELGLGIGLFQAGTNLFLNGMVLGTMFLGGHLMSTGQMSAGDVMAFLVNAQTVQRSVAQLSLLFGSVVKGISAGGRVFEYINKKPAMDTSGTKYIKYHEFHGDIEFKNVTFAYPTRPEAVILKDFNLKIPAGKTVAIVGTSGNGKSTIAALLERFYDVNEGQVSIDGVVLREFDGKWLRGRALGLIGQEPVLFATTVRENIRYGRPEATDAEVMQAATIANAHEFIQSFPEGYDTLVGERGMALSGGQKQRVAIARAVLKNPPVIILDEATSALDSASEKVVQKALETAAKGRTVLVIAHRLSTVINADLIVVLNKGKIVEMGNHEQLKKLKGFYWNLIKQQDQDLEQSR
Summary
Uniprot
H9J3Q5
A0A2H4KAW5
A0A3S2NW93
A0A2A4IWJ2
A0A2H1VIQ3
A0A194QIC9
+ More
A0A212FNN3 A0A0F6PQJ7 A0A067RAL4 A0A2J7PBG9 A0A2J7PBG0 A0A1B6CUH5 A0A1B6CLY8 A0A1B6CTZ2 A0A2R4FX95 D6WUA9 A0A0U9HU05 A0A1Y1LRP3 N6TUE5 U4UMX0 A0A158V4T9 A0A1B6GLK0 J9K9R9 A0A0B7AEV3 A0A0A7ARK3 A0A3R5SLD7 A0A1B6F4U7 A0A0P4W589 A0A1Y1LT88 A0A0P5GCP8 A0A0P5KKT0 K1PYF9 A0A2S2Q8Q1 A0A0P6D6N1 A0A0P5K7Z4 A0A0P4WAB7 A0A0P6GLK6 A0A0N8C578 A0A087ZV79 A0A0N8E5S6 A0A0N8E4R5 A0A2A3E5A5 A0A0B7AEF4 A0A0P5B6U9 A0A0P5DXX1 A0A0P5PYE4 A0A0P5ZW23 A0A0P5V6Z7 A0A0N8A1N3 A0A0P4ZWB9 A0A0P5V9Y6 A0A210Q3D9 R7U8L9 A0A0C9PXR9 A0A0P5DGJ4 V4A1S9 A0A2P2I6D0 A0A0K2SXV1 A0A0N7ZU64 A0A0P5MB61 A0A158NCG1 A0A195F9Z0 A0A2C9JDK1 A0A195BYA7 A0A131XA52 A0A026X184 F4WX04 A0A151JQT8 A0A0P6GI06 A0A023GND3 A0A0C9SCL6 A0A0P5VBC3 A0A0P6G9S4 A0A131YYF2 C3Z387 A0A224YI55 A0A293MRA2 A0A1E1X4U5 A0A0P5YPA6 A0A3S1BNR6 V5ID80 A0A2R5LMG0 V5HPY8 E9GHP1 A0A023F3L3 A0A1D1W6S4 A0A1W5ACC0 A0A0P4XS94 A0A224XGZ5 A0A0L7R1D0 A0A3Q2YPF0 E1ZWB0 A0A0J7KRR7 A0A1A8B5Q2 T1HNG3 W5MSG2 A0A1W5AKT8 A0A1Z5KTX6 A0A151WMT6 A0A2I4CZ49
A0A212FNN3 A0A0F6PQJ7 A0A067RAL4 A0A2J7PBG9 A0A2J7PBG0 A0A1B6CUH5 A0A1B6CLY8 A0A1B6CTZ2 A0A2R4FX95 D6WUA9 A0A0U9HU05 A0A1Y1LRP3 N6TUE5 U4UMX0 A0A158V4T9 A0A1B6GLK0 J9K9R9 A0A0B7AEV3 A0A0A7ARK3 A0A3R5SLD7 A0A1B6F4U7 A0A0P4W589 A0A1Y1LT88 A0A0P5GCP8 A0A0P5KKT0 K1PYF9 A0A2S2Q8Q1 A0A0P6D6N1 A0A0P5K7Z4 A0A0P4WAB7 A0A0P6GLK6 A0A0N8C578 A0A087ZV79 A0A0N8E5S6 A0A0N8E4R5 A0A2A3E5A5 A0A0B7AEF4 A0A0P5B6U9 A0A0P5DXX1 A0A0P5PYE4 A0A0P5ZW23 A0A0P5V6Z7 A0A0N8A1N3 A0A0P4ZWB9 A0A0P5V9Y6 A0A210Q3D9 R7U8L9 A0A0C9PXR9 A0A0P5DGJ4 V4A1S9 A0A2P2I6D0 A0A0K2SXV1 A0A0N7ZU64 A0A0P5MB61 A0A158NCG1 A0A195F9Z0 A0A2C9JDK1 A0A195BYA7 A0A131XA52 A0A026X184 F4WX04 A0A151JQT8 A0A0P6GI06 A0A023GND3 A0A0C9SCL6 A0A0P5VBC3 A0A0P6G9S4 A0A131YYF2 C3Z387 A0A224YI55 A0A293MRA2 A0A1E1X4U5 A0A0P5YPA6 A0A3S1BNR6 V5ID80 A0A2R5LMG0 V5HPY8 E9GHP1 A0A023F3L3 A0A1D1W6S4 A0A1W5ACC0 A0A0P4XS94 A0A224XGZ5 A0A0L7R1D0 A0A3Q2YPF0 E1ZWB0 A0A0J7KRR7 A0A1A8B5Q2 T1HNG3 W5MSG2 A0A1W5AKT8 A0A1Z5KTX6 A0A151WMT6 A0A2I4CZ49
Pubmed
EMBL
BABH01021768
BABH01021769
BABH01021770
KY484776
ASS36018.1
RSAL01000136
+ More
RVE46215.1 NWSH01005627 PCG64091.1 ODYU01002790 SOQ40720.1 KQ459185 KPJ03206.1 AGBW02005277 OWR55300.1 KM244037 AKC42330.1 KK852581 KDR20884.1 NEVH01027097 PNF13674.1 PNF13673.1 GEDC01029072 GEDC01020109 JAS08226.1 JAS17189.1 GEDC01022920 JAS14378.1 GEDC01020493 JAS16805.1 MF034755 AVT42121.1 KQ971352 EFA06793.1 GARF01000064 JAC88893.1 GEZM01050589 JAV75508.1 APGK01054091 KB741247 ENN72006.1 KB632281 ERL91345.1 KF828783 AIN44100.1 GECZ01006481 JAS63288.1 ABLF02020174 HACG01032302 CEK79167.1 KF906272 AHK05637.1 MH172497 QAA95904.1 GECZ01024876 JAS44893.1 GDRN01086554 JAI61163.1 GEZM01050588 JAV75510.1 GDIQ01246524 JAK05201.1 GDIQ01187855 JAK63870.1 JH817517 EKC24099.1 GGMS01004906 MBY74109.1 GDIQ01083296 JAN11441.1 GDIQ01194212 JAK57513.1 GDRN01086559 GDRN01086558 JAI61162.1 GDIQ01171845 GDIQ01073149 GDIQ01031861 JAN62876.1 GDIQ01113635 JAL38091.1 GDIQ01059060 GDIQ01056415 JAN35677.1 GDIQ01061948 JAN32789.1 KZ288362 PBC26943.1 HACG01032303 CEK79168.1 GDIP01188669 JAJ34733.1 GDIP01150134 JAJ73268.1 GDIQ01121711 JAL30015.1 GDIP01078783 GDIP01038237 JAM65478.1 GDIP01119433 JAL84281.1 GDIP01191273 JAJ32129.1 GDIP01220622 JAJ02780.1 GDIP01102430 JAM01285.1 NEDP02005138 OWF43263.1 AMQN01008834 KB304051 ELU02451.1 GBYB01006273 JAG76040.1 GDIP01156683 JAJ66719.1 KB201304 ESO97788.1 IACF01003971 LAB69574.1 HACA01000740 CDW18101.1 GDIP01212225 JAJ11177.1 GDIQ01166759 JAK84966.1 ADTU01011876 KQ981727 KYN37032.1 KQ976396 KYM92923.1 GEFH01004862 JAP63719.1 KK107034 QOIP01000008 EZA62047.1 RLU19719.1 GL888417 EGI61247.1 KQ978619 KYN29758.1 GDIQ01052802 JAN41935.1 GBBM01000062 JAC35356.1 GBZX01001253 JAG91487.1 GDIP01102429 JAM01286.1 GDIQ01036568 JAN58169.1 GEDV01004975 JAP83582.1 GG666576 EEN52931.1 GFPF01006190 MAA17336.1 GFWV01023450 MAA48177.1 GFAC01004913 JAT94275.1 GDIP01055045 JAM48670.1 RQTK01000182 RUS85087.1 GANP01012402 JAB72066.1 GGLE01006600 MBY10726.1 GANP01006641 JAB77827.1 GL732545 EFX81018.1 GBBI01003253 JAC15459.1 BDGG01000020 GAV09120.1 GDIP01238778 LRGB01002580 JAI84623.1 KZS06982.1 GFTR01007338 JAW09088.1 KQ414668 KOC64653.1 GL434791 EFN74514.1 LBMM01003975 KMQ92934.1 HADY01024089 SBP62574.1 ACPB03007695 ACPB03007696 AHAT01010188 AHAT01010189 GFJQ02008418 JAV98551.1 KQ982934 KYQ49176.1
RVE46215.1 NWSH01005627 PCG64091.1 ODYU01002790 SOQ40720.1 KQ459185 KPJ03206.1 AGBW02005277 OWR55300.1 KM244037 AKC42330.1 KK852581 KDR20884.1 NEVH01027097 PNF13674.1 PNF13673.1 GEDC01029072 GEDC01020109 JAS08226.1 JAS17189.1 GEDC01022920 JAS14378.1 GEDC01020493 JAS16805.1 MF034755 AVT42121.1 KQ971352 EFA06793.1 GARF01000064 JAC88893.1 GEZM01050589 JAV75508.1 APGK01054091 KB741247 ENN72006.1 KB632281 ERL91345.1 KF828783 AIN44100.1 GECZ01006481 JAS63288.1 ABLF02020174 HACG01032302 CEK79167.1 KF906272 AHK05637.1 MH172497 QAA95904.1 GECZ01024876 JAS44893.1 GDRN01086554 JAI61163.1 GEZM01050588 JAV75510.1 GDIQ01246524 JAK05201.1 GDIQ01187855 JAK63870.1 JH817517 EKC24099.1 GGMS01004906 MBY74109.1 GDIQ01083296 JAN11441.1 GDIQ01194212 JAK57513.1 GDRN01086559 GDRN01086558 JAI61162.1 GDIQ01171845 GDIQ01073149 GDIQ01031861 JAN62876.1 GDIQ01113635 JAL38091.1 GDIQ01059060 GDIQ01056415 JAN35677.1 GDIQ01061948 JAN32789.1 KZ288362 PBC26943.1 HACG01032303 CEK79168.1 GDIP01188669 JAJ34733.1 GDIP01150134 JAJ73268.1 GDIQ01121711 JAL30015.1 GDIP01078783 GDIP01038237 JAM65478.1 GDIP01119433 JAL84281.1 GDIP01191273 JAJ32129.1 GDIP01220622 JAJ02780.1 GDIP01102430 JAM01285.1 NEDP02005138 OWF43263.1 AMQN01008834 KB304051 ELU02451.1 GBYB01006273 JAG76040.1 GDIP01156683 JAJ66719.1 KB201304 ESO97788.1 IACF01003971 LAB69574.1 HACA01000740 CDW18101.1 GDIP01212225 JAJ11177.1 GDIQ01166759 JAK84966.1 ADTU01011876 KQ981727 KYN37032.1 KQ976396 KYM92923.1 GEFH01004862 JAP63719.1 KK107034 QOIP01000008 EZA62047.1 RLU19719.1 GL888417 EGI61247.1 KQ978619 KYN29758.1 GDIQ01052802 JAN41935.1 GBBM01000062 JAC35356.1 GBZX01001253 JAG91487.1 GDIP01102429 JAM01286.1 GDIQ01036568 JAN58169.1 GEDV01004975 JAP83582.1 GG666576 EEN52931.1 GFPF01006190 MAA17336.1 GFWV01023450 MAA48177.1 GFAC01004913 JAT94275.1 GDIP01055045 JAM48670.1 RQTK01000182 RUS85087.1 GANP01012402 JAB72066.1 GGLE01006600 MBY10726.1 GANP01006641 JAB77827.1 GL732545 EFX81018.1 GBBI01003253 JAC15459.1 BDGG01000020 GAV09120.1 GDIP01238778 LRGB01002580 JAI84623.1 KZS06982.1 GFTR01007338 JAW09088.1 KQ414668 KOC64653.1 GL434791 EFN74514.1 LBMM01003975 KMQ92934.1 HADY01024089 SBP62574.1 ACPB03007695 ACPB03007696 AHAT01010188 AHAT01010189 GFJQ02008418 JAV98551.1 KQ982934 KYQ49176.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000027135
+ More
UP000235965 UP000007266 UP000019118 UP000030742 UP000007819 UP000005408 UP000005203 UP000242457 UP000242188 UP000014760 UP000030746 UP000005205 UP000078541 UP000076420 UP000078540 UP000053097 UP000279307 UP000007755 UP000078492 UP000001554 UP000271974 UP000000305 UP000186922 UP000192224 UP000076858 UP000053825 UP000264820 UP000000311 UP000036403 UP000015103 UP000018468 UP000075809 UP000192220
UP000235965 UP000007266 UP000019118 UP000030742 UP000007819 UP000005408 UP000005203 UP000242457 UP000242188 UP000014760 UP000030746 UP000005205 UP000078541 UP000076420 UP000078540 UP000053097 UP000279307 UP000007755 UP000078492 UP000001554 UP000271974 UP000000305 UP000186922 UP000192224 UP000076858 UP000053825 UP000264820 UP000000311 UP000036403 UP000015103 UP000018468 UP000075809 UP000192220
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J3Q5
A0A2H4KAW5
A0A3S2NW93
A0A2A4IWJ2
A0A2H1VIQ3
A0A194QIC9
+ More
A0A212FNN3 A0A0F6PQJ7 A0A067RAL4 A0A2J7PBG9 A0A2J7PBG0 A0A1B6CUH5 A0A1B6CLY8 A0A1B6CTZ2 A0A2R4FX95 D6WUA9 A0A0U9HU05 A0A1Y1LRP3 N6TUE5 U4UMX0 A0A158V4T9 A0A1B6GLK0 J9K9R9 A0A0B7AEV3 A0A0A7ARK3 A0A3R5SLD7 A0A1B6F4U7 A0A0P4W589 A0A1Y1LT88 A0A0P5GCP8 A0A0P5KKT0 K1PYF9 A0A2S2Q8Q1 A0A0P6D6N1 A0A0P5K7Z4 A0A0P4WAB7 A0A0P6GLK6 A0A0N8C578 A0A087ZV79 A0A0N8E5S6 A0A0N8E4R5 A0A2A3E5A5 A0A0B7AEF4 A0A0P5B6U9 A0A0P5DXX1 A0A0P5PYE4 A0A0P5ZW23 A0A0P5V6Z7 A0A0N8A1N3 A0A0P4ZWB9 A0A0P5V9Y6 A0A210Q3D9 R7U8L9 A0A0C9PXR9 A0A0P5DGJ4 V4A1S9 A0A2P2I6D0 A0A0K2SXV1 A0A0N7ZU64 A0A0P5MB61 A0A158NCG1 A0A195F9Z0 A0A2C9JDK1 A0A195BYA7 A0A131XA52 A0A026X184 F4WX04 A0A151JQT8 A0A0P6GI06 A0A023GND3 A0A0C9SCL6 A0A0P5VBC3 A0A0P6G9S4 A0A131YYF2 C3Z387 A0A224YI55 A0A293MRA2 A0A1E1X4U5 A0A0P5YPA6 A0A3S1BNR6 V5ID80 A0A2R5LMG0 V5HPY8 E9GHP1 A0A023F3L3 A0A1D1W6S4 A0A1W5ACC0 A0A0P4XS94 A0A224XGZ5 A0A0L7R1D0 A0A3Q2YPF0 E1ZWB0 A0A0J7KRR7 A0A1A8B5Q2 T1HNG3 W5MSG2 A0A1W5AKT8 A0A1Z5KTX6 A0A151WMT6 A0A2I4CZ49
A0A212FNN3 A0A0F6PQJ7 A0A067RAL4 A0A2J7PBG9 A0A2J7PBG0 A0A1B6CUH5 A0A1B6CLY8 A0A1B6CTZ2 A0A2R4FX95 D6WUA9 A0A0U9HU05 A0A1Y1LRP3 N6TUE5 U4UMX0 A0A158V4T9 A0A1B6GLK0 J9K9R9 A0A0B7AEV3 A0A0A7ARK3 A0A3R5SLD7 A0A1B6F4U7 A0A0P4W589 A0A1Y1LT88 A0A0P5GCP8 A0A0P5KKT0 K1PYF9 A0A2S2Q8Q1 A0A0P6D6N1 A0A0P5K7Z4 A0A0P4WAB7 A0A0P6GLK6 A0A0N8C578 A0A087ZV79 A0A0N8E5S6 A0A0N8E4R5 A0A2A3E5A5 A0A0B7AEF4 A0A0P5B6U9 A0A0P5DXX1 A0A0P5PYE4 A0A0P5ZW23 A0A0P5V6Z7 A0A0N8A1N3 A0A0P4ZWB9 A0A0P5V9Y6 A0A210Q3D9 R7U8L9 A0A0C9PXR9 A0A0P5DGJ4 V4A1S9 A0A2P2I6D0 A0A0K2SXV1 A0A0N7ZU64 A0A0P5MB61 A0A158NCG1 A0A195F9Z0 A0A2C9JDK1 A0A195BYA7 A0A131XA52 A0A026X184 F4WX04 A0A151JQT8 A0A0P6GI06 A0A023GND3 A0A0C9SCL6 A0A0P5VBC3 A0A0P6G9S4 A0A131YYF2 C3Z387 A0A224YI55 A0A293MRA2 A0A1E1X4U5 A0A0P5YPA6 A0A3S1BNR6 V5ID80 A0A2R5LMG0 V5HPY8 E9GHP1 A0A023F3L3 A0A1D1W6S4 A0A1W5ACC0 A0A0P4XS94 A0A224XGZ5 A0A0L7R1D0 A0A3Q2YPF0 E1ZWB0 A0A0J7KRR7 A0A1A8B5Q2 T1HNG3 W5MSG2 A0A1W5AKT8 A0A1Z5KTX6 A0A151WMT6 A0A2I4CZ49
PDB
5OCH
E-value=2.80964e-116,
Score=1070
Ontologies
GO
PANTHER
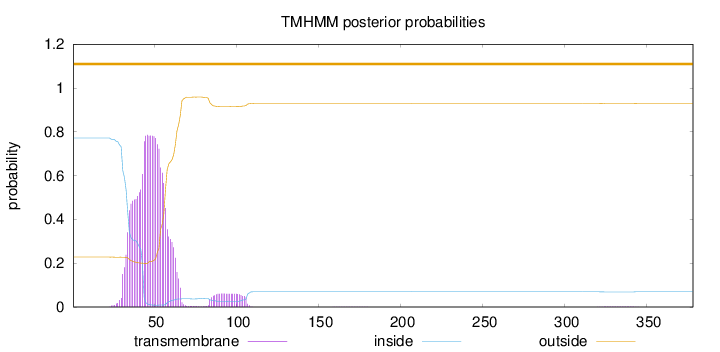
Topology
Length:
378
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
19.15287
Exp number, first 60 AAs:
16.5308
Total prob of N-in:
0.77187
POSSIBLE N-term signal
sequence
outside
1 - 378
Population Genetic Test Statistics
Pi
24.279653
Theta
21.709145
Tajima's D
-0.721685
CLR
0.932956
CSRT
0.186390680465977
Interpretation
Uncertain
