Pre Gene Modal
BGIBMGA004142
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_B_member_8?_mitochondrial-like?_partial_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 2.364
Sequence
CDS
ATGTGGTATATGTTACAGAATCACTTATTTTGTCTCACAAGACAAAGGTTCCTGGCTTCAAAAAGTAATAAATACTTCACTAATATAGTAAAAAACAAGCCATCAGACGCTTTAAATCGATATATATCACAGTTAAAGACTCGTGGAAAAAGTTCAAAGTCTGGAGAATCATTGCCTTGCAGTCCATGGAAATTGTGTACAGGAGTTAGTTTAGCTATTGGCGCTCGGTATTTTCTATATAGGAGTCCAGTATTTTGTAAAGCAGTTCAGACTTCGAGAGTAATACAGAGAAGGCCGAATTTCCAAGAGGATACCGCTAAATTTGATTGGGAAAAATTATTGAACTATTTAAAACCTCATAAATGGCTCCTTATAGCTGCTGTTGGTTCTGCGTTAGCTGTGGCATTCCTAAATGTGTACATCCCAGCTATGCTCGGTGTCATCGTGAATGTTCTAGCAGGAATAAGGAATAATCCGGCTGCTGATTTCATTGACGAGATCAAGATGCCGGCATTTAAATTGATTTCATTGTATGTTGGACAGGCAGCTTTTACGTTTTTCTACATTCACTTGCTGTCACAAGTGGGAGAAAAAGTGGCCACTCAAATGAAACAAGATCTATTTGTATCTATTCTAAGACAGGATATAGAATTTTTTGATAAAGAAAGAACTGGGGAATTAGTTAATAGGTTAACAGTGGACGTACAAGACTTCAAAAGCTCTTTCAAACAGACCATATCGGGAGGTCTTAGAGCTATTACGCAGGTGGTGGGTTCTGCGTTGTTATCGGAGGCACCTTCATTGGTTCGCTGTTGA
Protein
MWYMLQNHLFCLTRQRFLASKSNKYFTNIVKNKPSDALNRYISQLKTRGKSSKSGESLPCSPWKLCTGVSLAIGARYFLYRSPVFCKAVQTSRVIQRRPNFQEDTAKFDWEKLLNYLKPHKWLLIAAVGSALAVAFLNVYIPAMLGVIVNVLAGIRNNPAADFIDEIKMPAFKLISLYVGQAAFTFFYIHLLSQVGEKVATQMKQDLFVSILRQDIEFFDKERTGELVNRLTVDVQDFKSSFKQTISGGLRAITQVVGSALLSEAPSLVRC
Summary
Uniprot
A0A2H1VPU5
A0A2A4IWJ2
A0A2H1VIQ3
A0A3S2NW93
A0A2H4KAW5
A0A212FNN3
+ More
A0A194QYA3 A0A194QIC9 A0A0P4ZX17 A0A0P5E543 A0A0P5U6Y3 A0A0P5ZV27 A0A0P5TDA1 A0A0P5JL62 A0A0P5S0L7 A0A0N8C578 A0A0N8E4R5 A0A0P5V9Y6 A0A0P5PYE4 A0A0N7ZU64 A0A0P5ZW23 A0A0P6GLK6 A0A0P5DXX1 A0A0P4XS94 A0A0P4ZWB9 A0A0P6G9S4 A0A0P5YPA6 A0A0P6GI06 A0A0P5TF32 A0A0T6B5Z7 A0A0P6I9H4 A0A0N8A1N3 A0A0P5V6Z7 A0A0N8E5S6 A0A2J7PBH4 A0A067RAL4 T1PE36 A0A2J7PBG0 A0A1I8NEI4 Q17I35 A0A0L0BQU8 W8AYD4 A0A0A1XEY9 A0A1Y1LRP3 A0A1Y1LT88 A0A034VH39 A0A2H4TEP5 B4JNJ4 A0A1I8PBT0 B4GU29 Q29J72 A0A084WKF5 A0A0P5ZJ47 A0A2P8YZ46 E9GHP1 B4L6K4 A0A182GAI1 A0A0Q9XF01 A0A0K8VQ28 Q9VYL5 A0A182F9U1 A0A0K8TXX5 W5JVP9 Q7Q5Q2 B3MRZ4 A0A182TV50 B4R3Y6 A0A182X8Q5 A0A182I122 A0A154PP01 A0A182LLE1 B3NXB4 A0A182ULQ3 A0A1S3KF87 A0A1Q3F9U7 B0WV49 A0A158V4T9 A0A336LPL2 A0A182J1F7 A0A182S9W1 A0A182NJ88 A0A182QCH4
A0A194QYA3 A0A194QIC9 A0A0P4ZX17 A0A0P5E543 A0A0P5U6Y3 A0A0P5ZV27 A0A0P5TDA1 A0A0P5JL62 A0A0P5S0L7 A0A0N8C578 A0A0N8E4R5 A0A0P5V9Y6 A0A0P5PYE4 A0A0N7ZU64 A0A0P5ZW23 A0A0P6GLK6 A0A0P5DXX1 A0A0P4XS94 A0A0P4ZWB9 A0A0P6G9S4 A0A0P5YPA6 A0A0P6GI06 A0A0P5TF32 A0A0T6B5Z7 A0A0P6I9H4 A0A0N8A1N3 A0A0P5V6Z7 A0A0N8E5S6 A0A2J7PBH4 A0A067RAL4 T1PE36 A0A2J7PBG0 A0A1I8NEI4 Q17I35 A0A0L0BQU8 W8AYD4 A0A0A1XEY9 A0A1Y1LRP3 A0A1Y1LT88 A0A034VH39 A0A2H4TEP5 B4JNJ4 A0A1I8PBT0 B4GU29 Q29J72 A0A084WKF5 A0A0P5ZJ47 A0A2P8YZ46 E9GHP1 B4L6K4 A0A182GAI1 A0A0Q9XF01 A0A0K8VQ28 Q9VYL5 A0A182F9U1 A0A0K8TXX5 W5JVP9 Q7Q5Q2 B3MRZ4 A0A182TV50 B4R3Y6 A0A182X8Q5 A0A182I122 A0A154PP01 A0A182LLE1 B3NXB4 A0A182ULQ3 A0A1S3KF87 A0A1Q3F9U7 B0WV49 A0A158V4T9 A0A336LPL2 A0A182J1F7 A0A182S9W1 A0A182NJ88 A0A182QCH4
Pubmed
22118469
26354079
24845553
25315136
17510324
26108605
+ More
24495485 25830018 28004739 25348373 29121518 17994087 15632085 24438588 29403074 21292972 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 12364791 14747013 17210077 20966253
24495485 25830018 28004739 25348373 29121518 17994087 15632085 24438588 29403074 21292972 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 12364791 14747013 17210077 20966253
EMBL
ODYU01003453
SOQ42254.1
NWSH01005627
PCG64091.1
ODYU01002790
SOQ40720.1
+ More
RSAL01000136 RVE46215.1 KY484776 ASS36018.1 AGBW02005277 OWR55300.1 KQ460949 KPJ10447.1 KQ459185 KPJ03206.1 GDIP01211222 JAJ12180.1 GDIP01151428 JAJ71974.1 GDIP01134189 JAL69525.1 GDIP01039397 JAM64318.1 GDIP01131691 JAL72023.1 GDIQ01201566 JAK50159.1 GDIQ01100649 JAL51077.1 GDIQ01113635 JAL38091.1 GDIQ01061948 JAN32789.1 GDIP01102430 JAM01285.1 GDIQ01121711 JAL30015.1 GDIP01212225 JAJ11177.1 GDIP01078783 GDIP01038237 JAM65478.1 GDIQ01171845 GDIQ01073149 GDIQ01031861 JAN62876.1 GDIP01150134 JAJ73268.1 GDIP01238778 LRGB01002580 JAI84623.1 KZS06982.1 GDIP01220622 JAJ02780.1 GDIQ01036568 JAN58169.1 GDIP01055045 JAM48670.1 GDIQ01052802 JAN41935.1 GDIP01132984 JAL70730.1 LJIG01009566 KRT82824.1 GDIQ01007723 JAN87014.1 GDIP01191273 JAJ32129.1 GDIP01119433 JAL84281.1 GDIQ01059060 GDIQ01056415 JAN35677.1 NEVH01027097 PNF13672.1 KK852581 KDR20884.1 KA646405 AFP61034.1 PNF13673.1 CH477243 EAT46324.1 JRES01001496 KNC22455.1 GAMC01015424 GAMC01015423 JAB91132.1 GBXI01005239 JAD09053.1 GEZM01050589 JAV75508.1 GEZM01050588 JAV75510.1 GAKP01017862 JAC41090.1 KY849626 ATY74510.1 CH916371 EDV92287.1 CH479191 EDW26112.1 CH379063 EAL32430.2 ATLV01024118 KE525349 KFB50699.1 GDIP01056484 JAM47231.1 PYGN01000276 PSN49512.1 GL732545 EFX81018.1 CH933812 EDW06000.1 JXUM01051212 KQ561679 KXJ77820.1 KRG07115.1 GDHF01011366 JAI40948.1 AE014298 AY118801 AAF48177.1 AAM50661.1 GDHF01033000 JAI19314.1 ADMH02000335 ETN66919.1 AAAB01008960 EAA11750.4 CH902622 EDV34549.1 CM000366 EDX17753.1 APCN01000023 KQ435007 KZC13611.1 CH954180 EDV47286.1 GFDL01010752 JAV24293.1 DS232118 EDS35382.1 KF828783 AIN44100.1 UFQS01000039 UFQT01000039 SSW98113.1 SSX18499.1 AXCN02000452
RSAL01000136 RVE46215.1 KY484776 ASS36018.1 AGBW02005277 OWR55300.1 KQ460949 KPJ10447.1 KQ459185 KPJ03206.1 GDIP01211222 JAJ12180.1 GDIP01151428 JAJ71974.1 GDIP01134189 JAL69525.1 GDIP01039397 JAM64318.1 GDIP01131691 JAL72023.1 GDIQ01201566 JAK50159.1 GDIQ01100649 JAL51077.1 GDIQ01113635 JAL38091.1 GDIQ01061948 JAN32789.1 GDIP01102430 JAM01285.1 GDIQ01121711 JAL30015.1 GDIP01212225 JAJ11177.1 GDIP01078783 GDIP01038237 JAM65478.1 GDIQ01171845 GDIQ01073149 GDIQ01031861 JAN62876.1 GDIP01150134 JAJ73268.1 GDIP01238778 LRGB01002580 JAI84623.1 KZS06982.1 GDIP01220622 JAJ02780.1 GDIQ01036568 JAN58169.1 GDIP01055045 JAM48670.1 GDIQ01052802 JAN41935.1 GDIP01132984 JAL70730.1 LJIG01009566 KRT82824.1 GDIQ01007723 JAN87014.1 GDIP01191273 JAJ32129.1 GDIP01119433 JAL84281.1 GDIQ01059060 GDIQ01056415 JAN35677.1 NEVH01027097 PNF13672.1 KK852581 KDR20884.1 KA646405 AFP61034.1 PNF13673.1 CH477243 EAT46324.1 JRES01001496 KNC22455.1 GAMC01015424 GAMC01015423 JAB91132.1 GBXI01005239 JAD09053.1 GEZM01050589 JAV75508.1 GEZM01050588 JAV75510.1 GAKP01017862 JAC41090.1 KY849626 ATY74510.1 CH916371 EDV92287.1 CH479191 EDW26112.1 CH379063 EAL32430.2 ATLV01024118 KE525349 KFB50699.1 GDIP01056484 JAM47231.1 PYGN01000276 PSN49512.1 GL732545 EFX81018.1 CH933812 EDW06000.1 JXUM01051212 KQ561679 KXJ77820.1 KRG07115.1 GDHF01011366 JAI40948.1 AE014298 AY118801 AAF48177.1 AAM50661.1 GDHF01033000 JAI19314.1 ADMH02000335 ETN66919.1 AAAB01008960 EAA11750.4 CH902622 EDV34549.1 CM000366 EDX17753.1 APCN01000023 KQ435007 KZC13611.1 CH954180 EDV47286.1 GFDL01010752 JAV24293.1 DS232118 EDS35382.1 KF828783 AIN44100.1 UFQS01000039 UFQT01000039 SSW98113.1 SSX18499.1 AXCN02000452
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000076858
+ More
UP000235965 UP000027135 UP000095301 UP000008820 UP000037069 UP000001070 UP000095300 UP000008744 UP000001819 UP000030765 UP000245037 UP000000305 UP000009192 UP000069940 UP000249989 UP000000803 UP000069272 UP000000673 UP000007062 UP000007801 UP000075902 UP000000304 UP000076407 UP000075840 UP000076502 UP000075882 UP000008711 UP000075903 UP000085678 UP000002320 UP000075880 UP000075901 UP000075884 UP000075886
UP000235965 UP000027135 UP000095301 UP000008820 UP000037069 UP000001070 UP000095300 UP000008744 UP000001819 UP000030765 UP000245037 UP000000305 UP000009192 UP000069940 UP000249989 UP000000803 UP000069272 UP000000673 UP000007062 UP000007801 UP000075902 UP000000304 UP000076407 UP000075840 UP000076502 UP000075882 UP000008711 UP000075903 UP000085678 UP000002320 UP000075880 UP000075901 UP000075884 UP000075886
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VPU5
A0A2A4IWJ2
A0A2H1VIQ3
A0A3S2NW93
A0A2H4KAW5
A0A212FNN3
+ More
A0A194QYA3 A0A194QIC9 A0A0P4ZX17 A0A0P5E543 A0A0P5U6Y3 A0A0P5ZV27 A0A0P5TDA1 A0A0P5JL62 A0A0P5S0L7 A0A0N8C578 A0A0N8E4R5 A0A0P5V9Y6 A0A0P5PYE4 A0A0N7ZU64 A0A0P5ZW23 A0A0P6GLK6 A0A0P5DXX1 A0A0P4XS94 A0A0P4ZWB9 A0A0P6G9S4 A0A0P5YPA6 A0A0P6GI06 A0A0P5TF32 A0A0T6B5Z7 A0A0P6I9H4 A0A0N8A1N3 A0A0P5V6Z7 A0A0N8E5S6 A0A2J7PBH4 A0A067RAL4 T1PE36 A0A2J7PBG0 A0A1I8NEI4 Q17I35 A0A0L0BQU8 W8AYD4 A0A0A1XEY9 A0A1Y1LRP3 A0A1Y1LT88 A0A034VH39 A0A2H4TEP5 B4JNJ4 A0A1I8PBT0 B4GU29 Q29J72 A0A084WKF5 A0A0P5ZJ47 A0A2P8YZ46 E9GHP1 B4L6K4 A0A182GAI1 A0A0Q9XF01 A0A0K8VQ28 Q9VYL5 A0A182F9U1 A0A0K8TXX5 W5JVP9 Q7Q5Q2 B3MRZ4 A0A182TV50 B4R3Y6 A0A182X8Q5 A0A182I122 A0A154PP01 A0A182LLE1 B3NXB4 A0A182ULQ3 A0A1S3KF87 A0A1Q3F9U7 B0WV49 A0A158V4T9 A0A336LPL2 A0A182J1F7 A0A182S9W1 A0A182NJ88 A0A182QCH4
A0A194QYA3 A0A194QIC9 A0A0P4ZX17 A0A0P5E543 A0A0P5U6Y3 A0A0P5ZV27 A0A0P5TDA1 A0A0P5JL62 A0A0P5S0L7 A0A0N8C578 A0A0N8E4R5 A0A0P5V9Y6 A0A0P5PYE4 A0A0N7ZU64 A0A0P5ZW23 A0A0P6GLK6 A0A0P5DXX1 A0A0P4XS94 A0A0P4ZWB9 A0A0P6G9S4 A0A0P5YPA6 A0A0P6GI06 A0A0P5TF32 A0A0T6B5Z7 A0A0P6I9H4 A0A0N8A1N3 A0A0P5V6Z7 A0A0N8E5S6 A0A2J7PBH4 A0A067RAL4 T1PE36 A0A2J7PBG0 A0A1I8NEI4 Q17I35 A0A0L0BQU8 W8AYD4 A0A0A1XEY9 A0A1Y1LRP3 A0A1Y1LT88 A0A034VH39 A0A2H4TEP5 B4JNJ4 A0A1I8PBT0 B4GU29 Q29J72 A0A084WKF5 A0A0P5ZJ47 A0A2P8YZ46 E9GHP1 B4L6K4 A0A182GAI1 A0A0Q9XF01 A0A0K8VQ28 Q9VYL5 A0A182F9U1 A0A0K8TXX5 W5JVP9 Q7Q5Q2 B3MRZ4 A0A182TV50 B4R3Y6 A0A182X8Q5 A0A182I122 A0A154PP01 A0A182LLE1 B3NXB4 A0A182ULQ3 A0A1S3KF87 A0A1Q3F9U7 B0WV49 A0A158V4T9 A0A336LPL2 A0A182J1F7 A0A182S9W1 A0A182NJ88 A0A182QCH4
PDB
5OCH
E-value=6.34583e-30,
Score=324
Ontologies
GO
PANTHER
Topology
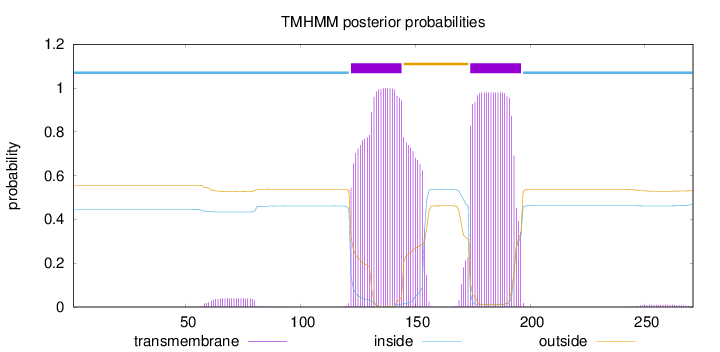
Length:
271
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
48.89515
Exp number, first 60 AAs:
0.06153
Total prob of N-in:
0.44503
inside
1 - 121
TMhelix
122 - 144
outside
145 - 173
TMhelix
174 - 196
inside
197 - 271
Population Genetic Test Statistics
Pi
26.751426
Theta
31.182308
Tajima's D
-0.704604
CLR
1.672803
CSRT
0.195590220488976
Interpretation
Uncertain
