Gene
KWMTBOMO11695
Pre Gene Modal
BGIBMGA003968
Annotation
PREDICTED:_putative_phosphatidate_phosphatase_[Papilio_xuthus]
Full name
Putative phosphatidate phosphatase
Alternative Name
Germ cell guidance factor
Phosphatidic acid phosphatase type 2
Protein wunen
Phosphatidic acid phosphatase type 2
Protein wunen
Location in the cell
PlasmaMembrane Reliability : 4.881
Sequence
CDS
ATGGCTGTTGATAGATTAATATGGAAAATTATTGTGGATTTCGTTGTGCTCGCGTGCGTTTCATTCCCCCTGCTTGCTCTGCTGCTGTGGGCGGAGCCCTATCACCGCGGCTACTTTGAAGATGACCTTTCTCTGCGACTGCCCTTTAAACAGCAGTCCATATCGGAAGGGCTCTTGGCTGGCGTCGGCTTCGCTTTCATTATTTTCACGGTTCTAATCACTGAGATAGTGCGTGACAGGCAAGGTAAGGGCATCGGCGGAAAGTTTCTGTCGGGGTCGCTGATACCGGGCTGGCTGTGGGAGACGTACTCGACCGTGGGAATCTTCACGTTTGGGGCCGCCTGTCAACAGCTCACAGCCAACCTCGCGAAGTACGTGATCGGGAGACTGCGCCCACATTTCTTCGATGTCTGCAGGCCGATCCCGGACGCCGGATCCTCGCAGAACGCCCTCGGCTACATCCAGGAGTACCGATGTAGCGGTGGCGATGAGGCTCTCTTGAAGGACATGCGCCTCTCGTTTCCGAGTGCACACGCAAGCTTCTCTATGTACTGTGCCATATTCTTTATCTTCTACATCCAAGTGAAAGGCAAGTGGCGGGGCTCGAAGCTCCTCCGTCACGGAGTCCAGTATGCCGTTGTGATAGCGGCCTGGTACGTGGGGCTCTCGAGGGTGGTGGAACATATGCACCACTGGAGCGACGTGGCGGCTGGTTTCGCCATAGGCGCCACATACGCTATGCTTATTTTCATCTTCGTTCTGAAGCCGAAGAAGTACGGCGCACCAGAGACGTGGCAGCAGCCGGTGCCCAGGAATACGCTTCCCAGGCCGGTACTGGCGCGGTGA
Protein
MAVDRLIWKIIVDFVVLACVSFPLLALLLWAEPYHRGYFEDDLSLRLPFKQQSISEGLLAGVGFAFIIFTVLITEIVRDRQGKGIGGKFLSGSLIPGWLWETYSTVGIFTFGAACQQLTANLAKYVIGRLRPHFFDVCRPIPDAGSSQNALGYIQEYRCSGGDEALLKDMRLSFPSAHASFSMYCAIFFIFYIQVKGKWRGSKLLRHGVQYAVVIAAWYVGLSRVVEHMHHWSDVAAGFAIGATYAMLIFIFVLKPKKYGAPETWQQPVPRNTLPRPVLAR
Summary
Description
Responsible for guiding the germ cells early in the process of migration from the lumen of the developing gut towards the overlying mesoderm, where the germ cells enter the gonads. May be involved in lipid metabolism.
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phosphate + H2O = a 1,2-diacyl-sn-glycerol + phosphate
Subunit
Homodimer. This complex seems not to be involved in substrate recognition, it may confer only structural or functional stability.
Similarity
Belongs to the PA-phosphatase related phosphoesterase family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Glycoprotein
Hydrolase
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Putative phosphatidate phosphatase
splice variant In isoform Short.
splice variant In isoform Short.
Uniprot
H9J381
A0A194R3N3
A0A194Q9B3
A0A2H1WI45
A0A194QE37
A0A2H1W730
+ More
A0A212FNI3 S4PY41 A0A212FNH6 A0A2A4JNX9 A0A3S2LX18 A0A1Y1MVF2 B4J876 B0VZ03 A0A1Q3FHC6 A0A1L8DY84 B4KQ09 B4MDV3 A0A1L8E725 A0A0Q9W0A7 Q172P3 A0A1L8DYT5 A0A0Q9XJA5 A0A0K8TTU9 A0A2J7R5L3 A0A1L8EIS0 A0A1L8EIW2 A0A1L8EIM8 A0A1L8E5Z5 W8BIL3 A0A2M4CJY6 A0A1L8E5W3 A0A2M4CIP2 A0A1J1ISA8 A0A0A1XTI2 A0A023EPH9 A0A2M4ARF8 A0A3B0J214 B4MPQ4 T1HKB2 A0A182HE45 A0A182QVZ1 D3TMP7 A0A1L8EJ03 A0A3B0IY91 A0A1L8EJ51 A0A1L8EIR9 A0A067QTP7 A0A1B6KNY2 A0A034VNU9 A0A0K8VEM0 B3MFK0 E0VQY9 A0A182FLB6 A0A034VMY9 A0A224XH03 A0A336MP79 B4GHS9 A0A0J9R9Z7 A0A0P9BY25 W8CAT4 B4QH88 A0A182JFU0 A0A0A9Y9L2 A0A0M4EKC2 A0A0K8S5D9 A0A0R1DMD4 U5EWL0 Q28ZG6 A0A0A9Y520 A0A084WDR5 N6WBT2 A0A2M3Z9K7 A0A1B6GLP9 B4HSS1 A0A0V0G785 C7LA91 B4P418 A0A0B4K7K7 Q9V576-2 A0A0K8S456 A0A146KZH3 Q9V576 B3N7H0 A0A0Q5WNR8 A0A0P5J5A0 A0A1W4VCA5 A0A0P5KF95 A0A0P5KUL5 A0A026WG83 A0A1B0AHS6 A0A1W4VBV9 E2BZ71 A0A0G3YL55 A0A0P5CUQ6 A0A0N8A367 E2ASD3 A0A182WM77
A0A212FNI3 S4PY41 A0A212FNH6 A0A2A4JNX9 A0A3S2LX18 A0A1Y1MVF2 B4J876 B0VZ03 A0A1Q3FHC6 A0A1L8DY84 B4KQ09 B4MDV3 A0A1L8E725 A0A0Q9W0A7 Q172P3 A0A1L8DYT5 A0A0Q9XJA5 A0A0K8TTU9 A0A2J7R5L3 A0A1L8EIS0 A0A1L8EIW2 A0A1L8EIM8 A0A1L8E5Z5 W8BIL3 A0A2M4CJY6 A0A1L8E5W3 A0A2M4CIP2 A0A1J1ISA8 A0A0A1XTI2 A0A023EPH9 A0A2M4ARF8 A0A3B0J214 B4MPQ4 T1HKB2 A0A182HE45 A0A182QVZ1 D3TMP7 A0A1L8EJ03 A0A3B0IY91 A0A1L8EJ51 A0A1L8EIR9 A0A067QTP7 A0A1B6KNY2 A0A034VNU9 A0A0K8VEM0 B3MFK0 E0VQY9 A0A182FLB6 A0A034VMY9 A0A224XH03 A0A336MP79 B4GHS9 A0A0J9R9Z7 A0A0P9BY25 W8CAT4 B4QH88 A0A182JFU0 A0A0A9Y9L2 A0A0M4EKC2 A0A0K8S5D9 A0A0R1DMD4 U5EWL0 Q28ZG6 A0A0A9Y520 A0A084WDR5 N6WBT2 A0A2M3Z9K7 A0A1B6GLP9 B4HSS1 A0A0V0G785 C7LA91 B4P418 A0A0B4K7K7 Q9V576-2 A0A0K8S456 A0A146KZH3 Q9V576 B3N7H0 A0A0Q5WNR8 A0A0P5J5A0 A0A1W4VCA5 A0A0P5KF95 A0A0P5KUL5 A0A026WG83 A0A1B0AHS6 A0A1W4VBV9 E2BZ71 A0A0G3YL55 A0A0P5CUQ6 A0A0N8A367 E2ASD3 A0A182WM77
EC Number
3.1.3.4
Pubmed
19121390
26354079
22118469
23622113
28004739
17994087
+ More
17510324 26369729 24495485 25830018 24945155 26483478 20353571 24845553 25348373 18057021 20566863 22936249 25401762 17550304 15632085 26823975 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8985246 10731138 12537569 12856002 14725715 24508170 30249741 20798317 25702953
17510324 26369729 24495485 25830018 24945155 26483478 20353571 24845553 25348373 18057021 20566863 22936249 25401762 17550304 15632085 26823975 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8985246 10731138 12537569 12856002 14725715 24508170 30249741 20798317 25702953
EMBL
BABH01021749
KQ460949
KPJ10451.1
KQ459252
KPJ02102.1
ODYU01008797
+ More
SOQ52707.1 KQ459185 KPJ03210.1 ODYU01006741 SOQ48868.1 AGBW02005277 OWR55304.1 GAIX01003948 JAA88612.1 OWR55302.1 NWSH01000947 PCG73418.1 RSAL01000136 RVE46217.1 GEZM01023368 JAV88425.1 CH916367 EDW01213.1 DS231813 EDS25757.1 GFDL01008088 JAV26957.1 GFDF01002710 JAV11374.1 CH933808 EDW08111.2 CH940662 EDW58718.1 GFDG01004345 JAV14454.1 KRF78429.1 CH477433 EAT41005.1 GFDF01002649 JAV11435.1 KRG03912.1 GDAI01000228 JAI17375.1 NEVH01006994 PNF36111.1 GFDG01000241 JAV18558.1 GFDG01000240 JAV18559.1 GFDG01000242 JAV18557.1 GFDG01004671 JAV14128.1 GAMC01005437 GAMC01005435 JAC01121.1 GGFL01001030 MBW65208.1 GFDG01004672 JAV14127.1 GGFL01001029 MBW65207.1 CVRI01000059 CRL03032.1 GBXI01000199 JAD14093.1 JXUM01023906 JXUM01023907 JXUM01023908 JXUM01023909 GAPW01002201 KQ560674 JAC11397.1 KXJ81330.1 GGFK01010064 MBW43385.1 OUUW01000001 SPP72963.1 CH963849 EDW74093.1 ACPB03006494 JXUM01035044 JXUM01035045 JXUM01035046 JXUM01035047 JXUM01035048 JXUM01035049 KQ561046 KXJ79901.1 AXCN02000966 EZ422699 ADD18975.1 GFDG01000175 JAV18624.1 SPP72964.1 GFDG01000176 JAV18623.1 GFDG01000244 JAV18555.1 KK853284 KDR08980.1 GEBQ01026846 JAT13131.1 GAKP01015462 JAC43490.1 GDHF01028491 GDHF01015052 JAI23823.1 JAI37262.1 CH902619 EDV36685.1 KPU76358.1 DS235442 EEB15795.1 GAKP01015460 JAC43492.1 GFTR01004661 JAW11765.1 UFQT01001933 SSX32172.1 CH479183 EDW36049.1 CM002911 KMY92509.1 KPU76359.1 GAMC01005436 JAC01120.1 CM000362 EDX06342.1 KMY92508.1 GBHO01017402 GBHO01017399 JAG26202.1 JAG26205.1 CP012524 ALC41817.1 GBRD01017872 JAG47955.1 CM000157 KRJ98495.1 GANO01001369 JAB58502.1 CM000071 EAL25647.2 GBHO01017401 GDHC01017279 GDHC01000397 JAG26203.1 JAQ01350.1 JAQ18232.1 ATLV01023054 KE525340 KFB48359.1 ENO01639.1 GGFM01004431 MBW25182.1 GECZ01006414 JAS63355.1 CH480816 EDW47098.1 GECL01002156 JAP03968.1 BT099641 ACU68950.1 EDW89501.1 AE013599 AFH07978.1 U73822 AF145595 BT001729 AY046533 GBRD01017873 JAG47954.1 GDHC01018237 JAQ00392.1 CH954177 EDV59375.1 KQS70778.1 GDIQ01210794 JAK40931.1 GDIQ01186573 JAK65152.1 GDIQ01186572 JAK65153.1 KK107231 QOIP01000009 EZA55072.1 RLU18933.1 GL451577 EFN78977.1 KM507105 AKM28429.1 GDIP01170944 JAJ52458.1 GDIP01187001 JAJ36401.1 GL442284 EFN63667.1
SOQ52707.1 KQ459185 KPJ03210.1 ODYU01006741 SOQ48868.1 AGBW02005277 OWR55304.1 GAIX01003948 JAA88612.1 OWR55302.1 NWSH01000947 PCG73418.1 RSAL01000136 RVE46217.1 GEZM01023368 JAV88425.1 CH916367 EDW01213.1 DS231813 EDS25757.1 GFDL01008088 JAV26957.1 GFDF01002710 JAV11374.1 CH933808 EDW08111.2 CH940662 EDW58718.1 GFDG01004345 JAV14454.1 KRF78429.1 CH477433 EAT41005.1 GFDF01002649 JAV11435.1 KRG03912.1 GDAI01000228 JAI17375.1 NEVH01006994 PNF36111.1 GFDG01000241 JAV18558.1 GFDG01000240 JAV18559.1 GFDG01000242 JAV18557.1 GFDG01004671 JAV14128.1 GAMC01005437 GAMC01005435 JAC01121.1 GGFL01001030 MBW65208.1 GFDG01004672 JAV14127.1 GGFL01001029 MBW65207.1 CVRI01000059 CRL03032.1 GBXI01000199 JAD14093.1 JXUM01023906 JXUM01023907 JXUM01023908 JXUM01023909 GAPW01002201 KQ560674 JAC11397.1 KXJ81330.1 GGFK01010064 MBW43385.1 OUUW01000001 SPP72963.1 CH963849 EDW74093.1 ACPB03006494 JXUM01035044 JXUM01035045 JXUM01035046 JXUM01035047 JXUM01035048 JXUM01035049 KQ561046 KXJ79901.1 AXCN02000966 EZ422699 ADD18975.1 GFDG01000175 JAV18624.1 SPP72964.1 GFDG01000176 JAV18623.1 GFDG01000244 JAV18555.1 KK853284 KDR08980.1 GEBQ01026846 JAT13131.1 GAKP01015462 JAC43490.1 GDHF01028491 GDHF01015052 JAI23823.1 JAI37262.1 CH902619 EDV36685.1 KPU76358.1 DS235442 EEB15795.1 GAKP01015460 JAC43492.1 GFTR01004661 JAW11765.1 UFQT01001933 SSX32172.1 CH479183 EDW36049.1 CM002911 KMY92509.1 KPU76359.1 GAMC01005436 JAC01120.1 CM000362 EDX06342.1 KMY92508.1 GBHO01017402 GBHO01017399 JAG26202.1 JAG26205.1 CP012524 ALC41817.1 GBRD01017872 JAG47955.1 CM000157 KRJ98495.1 GANO01001369 JAB58502.1 CM000071 EAL25647.2 GBHO01017401 GDHC01017279 GDHC01000397 JAG26203.1 JAQ01350.1 JAQ18232.1 ATLV01023054 KE525340 KFB48359.1 ENO01639.1 GGFM01004431 MBW25182.1 GECZ01006414 JAS63355.1 CH480816 EDW47098.1 GECL01002156 JAP03968.1 BT099641 ACU68950.1 EDW89501.1 AE013599 AFH07978.1 U73822 AF145595 BT001729 AY046533 GBRD01017873 JAG47954.1 GDHC01018237 JAQ00392.1 CH954177 EDV59375.1 KQS70778.1 GDIQ01210794 JAK40931.1 GDIQ01186573 JAK65152.1 GDIQ01186572 JAK65153.1 KK107231 QOIP01000009 EZA55072.1 RLU18933.1 GL451577 EFN78977.1 KM507105 AKM28429.1 GDIP01170944 JAJ52458.1 GDIP01187001 JAJ36401.1 GL442284 EFN63667.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000283053
+ More
UP000001070 UP000002320 UP000009192 UP000008792 UP000008820 UP000235965 UP000183832 UP000069940 UP000249989 UP000268350 UP000007798 UP000015103 UP000075886 UP000027135 UP000007801 UP000009046 UP000069272 UP000008744 UP000000304 UP000075880 UP000092553 UP000002282 UP000001819 UP000030765 UP000001292 UP000000803 UP000008711 UP000192221 UP000053097 UP000279307 UP000092445 UP000008237 UP000000311 UP000075920
UP000001070 UP000002320 UP000009192 UP000008792 UP000008820 UP000235965 UP000183832 UP000069940 UP000249989 UP000268350 UP000007798 UP000015103 UP000075886 UP000027135 UP000007801 UP000009046 UP000069272 UP000008744 UP000000304 UP000075880 UP000092553 UP000002282 UP000001819 UP000030765 UP000001292 UP000000803 UP000008711 UP000192221 UP000053097 UP000279307 UP000092445 UP000008237 UP000000311 UP000075920
Pfam
PF01569 PAP2
SUPFAM
SSF48317
SSF48317
ProteinModelPortal
H9J381
A0A194R3N3
A0A194Q9B3
A0A2H1WI45
A0A194QE37
A0A2H1W730
+ More
A0A212FNI3 S4PY41 A0A212FNH6 A0A2A4JNX9 A0A3S2LX18 A0A1Y1MVF2 B4J876 B0VZ03 A0A1Q3FHC6 A0A1L8DY84 B4KQ09 B4MDV3 A0A1L8E725 A0A0Q9W0A7 Q172P3 A0A1L8DYT5 A0A0Q9XJA5 A0A0K8TTU9 A0A2J7R5L3 A0A1L8EIS0 A0A1L8EIW2 A0A1L8EIM8 A0A1L8E5Z5 W8BIL3 A0A2M4CJY6 A0A1L8E5W3 A0A2M4CIP2 A0A1J1ISA8 A0A0A1XTI2 A0A023EPH9 A0A2M4ARF8 A0A3B0J214 B4MPQ4 T1HKB2 A0A182HE45 A0A182QVZ1 D3TMP7 A0A1L8EJ03 A0A3B0IY91 A0A1L8EJ51 A0A1L8EIR9 A0A067QTP7 A0A1B6KNY2 A0A034VNU9 A0A0K8VEM0 B3MFK0 E0VQY9 A0A182FLB6 A0A034VMY9 A0A224XH03 A0A336MP79 B4GHS9 A0A0J9R9Z7 A0A0P9BY25 W8CAT4 B4QH88 A0A182JFU0 A0A0A9Y9L2 A0A0M4EKC2 A0A0K8S5D9 A0A0R1DMD4 U5EWL0 Q28ZG6 A0A0A9Y520 A0A084WDR5 N6WBT2 A0A2M3Z9K7 A0A1B6GLP9 B4HSS1 A0A0V0G785 C7LA91 B4P418 A0A0B4K7K7 Q9V576-2 A0A0K8S456 A0A146KZH3 Q9V576 B3N7H0 A0A0Q5WNR8 A0A0P5J5A0 A0A1W4VCA5 A0A0P5KF95 A0A0P5KUL5 A0A026WG83 A0A1B0AHS6 A0A1W4VBV9 E2BZ71 A0A0G3YL55 A0A0P5CUQ6 A0A0N8A367 E2ASD3 A0A182WM77
A0A212FNI3 S4PY41 A0A212FNH6 A0A2A4JNX9 A0A3S2LX18 A0A1Y1MVF2 B4J876 B0VZ03 A0A1Q3FHC6 A0A1L8DY84 B4KQ09 B4MDV3 A0A1L8E725 A0A0Q9W0A7 Q172P3 A0A1L8DYT5 A0A0Q9XJA5 A0A0K8TTU9 A0A2J7R5L3 A0A1L8EIS0 A0A1L8EIW2 A0A1L8EIM8 A0A1L8E5Z5 W8BIL3 A0A2M4CJY6 A0A1L8E5W3 A0A2M4CIP2 A0A1J1ISA8 A0A0A1XTI2 A0A023EPH9 A0A2M4ARF8 A0A3B0J214 B4MPQ4 T1HKB2 A0A182HE45 A0A182QVZ1 D3TMP7 A0A1L8EJ03 A0A3B0IY91 A0A1L8EJ51 A0A1L8EIR9 A0A067QTP7 A0A1B6KNY2 A0A034VNU9 A0A0K8VEM0 B3MFK0 E0VQY9 A0A182FLB6 A0A034VMY9 A0A224XH03 A0A336MP79 B4GHS9 A0A0J9R9Z7 A0A0P9BY25 W8CAT4 B4QH88 A0A182JFU0 A0A0A9Y9L2 A0A0M4EKC2 A0A0K8S5D9 A0A0R1DMD4 U5EWL0 Q28ZG6 A0A0A9Y520 A0A084WDR5 N6WBT2 A0A2M3Z9K7 A0A1B6GLP9 B4HSS1 A0A0V0G785 C7LA91 B4P418 A0A0B4K7K7 Q9V576-2 A0A0K8S456 A0A146KZH3 Q9V576 B3N7H0 A0A0Q5WNR8 A0A0P5J5A0 A0A1W4VCA5 A0A0P5KF95 A0A0P5KUL5 A0A026WG83 A0A1B0AHS6 A0A1W4VBV9 E2BZ71 A0A0G3YL55 A0A0P5CUQ6 A0A0N8A367 E2ASD3 A0A182WM77
Ontologies
PATHWAY
00561
Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00564 Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00600 Sphingolipid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00564 Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00600 Sphingolipid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Membrane
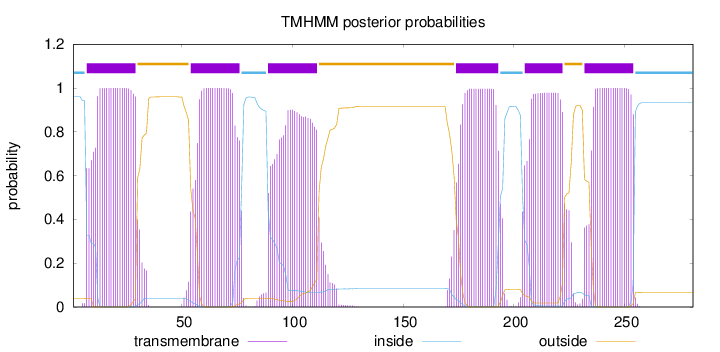
Length:
281
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
124.26041
Exp number, first 60 AAs:
28.06507
Total prob of N-in:
0.95978
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 53
TMhelix
54 - 76
inside
77 - 88
TMhelix
89 - 111
outside
112 - 173
TMhelix
174 - 193
inside
194 - 204
TMhelix
205 - 222
outside
223 - 231
TMhelix
232 - 254
inside
255 - 281
Population Genetic Test Statistics
Pi
19.340456
Theta
18.14472
Tajima's D
-1.055387
CLR
0.803256
CSRT
0.128943552822359
Interpretation
Uncertain
