Gene
KWMTBOMO11694
Pre Gene Modal
BGIBMGA003969
Annotation
PREDICTED:_putative_phosphatidate_phosphatase_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.874
Sequence
CDS
ATGTCTAGAGACGATACGTCAGTTCACATGCTAAGGAGTTTCGTTGGCGACATATTCCTATTGGTCGGACTATTGGTGTGTAATCTCTCATTAGAGAAGCTATGGGTTCAAACGCATCGGGGTTTCTTCTGTGGCGACGAGAGCTTAATGCTCCCATACAGAGACGACACAGTTACGGAATTAGTGCTGAGAATAATTGGAATAATCCCGCCTAGTATATTAATTTTAATCTTCGAGTGGACTCTTCTGCGCAAGGAACGCAGCGCAAGATGTTTCTCCATACCCGTCCCGTGGTGGGTGAGAAGTTCCTTTGGCATTCTTATGTCGTTCTCGGTCGGGTTTTGCTTCAATGAAATATTTACGAACATGGGAAAGAAAGTCATTGGAAGACTTAGACCACATTTCTTTGATTTATGCAAGCCTTCAGTGGACTGCAGTTTGCCAACTGACTTCGGTCGGTACATCGCTCCGAATGAGTACACATGCACAGCCGAAACTCCACTCTTGACTGATAATTTAAGAAAATCATTCCCCAGCGGACATTCTTCGAGTATCGCCTACACTGCGATTTTTATAGCTATATACTTAGAAAGGCGCTCAGTGTGGAAGAACCACAGGATCTTTCGCCACACTATTCAGTTGGCATTTTTACTGATGAGTTGGTACGTCGCCTTATCAAGAATAACAGACTTTAAGCATCACTGGAGCGATGTTCTATGTGGATATGCTCTGGGAATGTTCATCGCTCTATTTATGGCTGTAAGAGGAACTGAGCTCATTGAGGCGAAGAAAAACAATTCATCACCAGCACACGAAGTTAACCTTAATCATCAAGAACTGCCTATTAGTGCGTGA
Protein
MSRDDTSVHMLRSFVGDIFLLVGLLVCNLSLEKLWVQTHRGFFCGDESLMLPYRDDTVTELVLRIIGIIPPSILILIFEWTLLRKERSARCFSIPVPWWVRSSFGILMSFSVGFCFNEIFTNMGKKVIGRLRPHFFDLCKPSVDCSLPTDFGRYIAPNEYTCTAETPLLTDNLRKSFPSGHSSSIAYTAIFIAIYLERRSVWKNHRIFRHTIQLAFLLMSWYVALSRITDFKHHWSDVLCGYALGMFIALFMAVRGTELIEAKKNNSSPAHEVNLNHQELPISA
Summary
Uniprot
A0A194QID4
A0A194QYA8
A0A2H1V758
I4DKW4
A0A3S2LGJ6
A0A1E1WNF3
+ More
A0A2A4J496 A0A212FNJ1 S4PXA1 A0A2H1UZR4 A0A212FNH6 A0A3S2LX18 A0A2A4JNX9 A0A232FAM8 A0A023EPH9 A0A182HE45 A0A1Q3FHC6 B0VZ03 E2ASD3 Q172P3 A0A0L7R4Y5 E2BZ71 A0A182YQY6 K7J4V2 A0A182QVZ1 A0A195FSE8 A0A182VKV9 A0A1J1ISA8 A0A182FLB6 A0A182RUT8 A0A0G3YL55 A0A195C2I9 A0A336MP79 Q7QAY1 A0A2M4CIP2 A0A2M4CJY6 A0A084WDR5 A0A182JFU0 A0A158NNZ1 A0A182WM77 A0A2M4ARF8 A0A0A9Y520 A0A2M3Z9K7 A0A151HZK1 A0A151X7U8 J9JYU5 A0A0K8S456 F4WBK7 E0VQY9 A0A026WG83 A0A067QTP7 A0A2H8TXM7 T1HKB2 A0A0K8TTU9 U5EWL0 A0A146KZH3 A0A1Z5LES4 A0A0A9Y9L2 A0A0M4EKC2 A0A2J7R5L3 A0A0K8S5D9 B4KQ09 A0A0V0G785 A0A1B6C395 A0A224XH03 B4MDV3 A0A3Q0IQY3 A0A1J1IU97 B4J876 A0A182NLS8 A0A1B6KNY2 A0A0Q9W0A7 A0A087UAT8 W8BIL3 A0A1B6GLP9 A0A0Q9XJA5 A0A1B6E443 A0A182PVZ8 A0A1W4V011 A0A293N3I0 D3TMP7 A0A0A1XTI2 W8CAT4 A0A3B0JYZ9 A0A1B6CQX8 A0A194R3N3 A0A1L8E725 A0A1B0AHS6 A0A2T7PIV9 B4MPQ4 A0A1L8EIS0 A0A1L8EIM8 A0A1L8EIW2 A0A0P8XP46 E9GJV9
A0A2A4J496 A0A212FNJ1 S4PXA1 A0A2H1UZR4 A0A212FNH6 A0A3S2LX18 A0A2A4JNX9 A0A232FAM8 A0A023EPH9 A0A182HE45 A0A1Q3FHC6 B0VZ03 E2ASD3 Q172P3 A0A0L7R4Y5 E2BZ71 A0A182YQY6 K7J4V2 A0A182QVZ1 A0A195FSE8 A0A182VKV9 A0A1J1ISA8 A0A182FLB6 A0A182RUT8 A0A0G3YL55 A0A195C2I9 A0A336MP79 Q7QAY1 A0A2M4CIP2 A0A2M4CJY6 A0A084WDR5 A0A182JFU0 A0A158NNZ1 A0A182WM77 A0A2M4ARF8 A0A0A9Y520 A0A2M3Z9K7 A0A151HZK1 A0A151X7U8 J9JYU5 A0A0K8S456 F4WBK7 E0VQY9 A0A026WG83 A0A067QTP7 A0A2H8TXM7 T1HKB2 A0A0K8TTU9 U5EWL0 A0A146KZH3 A0A1Z5LES4 A0A0A9Y9L2 A0A0M4EKC2 A0A2J7R5L3 A0A0K8S5D9 B4KQ09 A0A0V0G785 A0A1B6C395 A0A224XH03 B4MDV3 A0A3Q0IQY3 A0A1J1IU97 B4J876 A0A182NLS8 A0A1B6KNY2 A0A0Q9W0A7 A0A087UAT8 W8BIL3 A0A1B6GLP9 A0A0Q9XJA5 A0A1B6E443 A0A182PVZ8 A0A1W4V011 A0A293N3I0 D3TMP7 A0A0A1XTI2 W8CAT4 A0A3B0JYZ9 A0A1B6CQX8 A0A194R3N3 A0A1L8E725 A0A1B0AHS6 A0A2T7PIV9 B4MPQ4 A0A1L8EIS0 A0A1L8EIM8 A0A1L8EIW2 A0A0P8XP46 E9GJV9
Pubmed
EMBL
KQ459185
KPJ03211.1
KQ460949
KPJ10452.1
ODYU01000841
SOQ36212.1
+ More
AK401932 BAM18554.1 RSAL01000136 RVE46219.1 GDQN01002683 GDQN01001775 JAT88371.1 JAT89279.1 NWSH01003423 PCG66334.1 AGBW02005277 OWR55305.1 GAIX01005398 JAA87162.1 ODYU01000021 SOQ34090.1 OWR55302.1 RVE46217.1 NWSH01000947 PCG73418.1 NNAY01000614 OXU27387.1 JXUM01023906 JXUM01023907 JXUM01023908 JXUM01023909 GAPW01002201 KQ560674 JAC11397.1 KXJ81330.1 JXUM01035044 JXUM01035045 JXUM01035046 JXUM01035047 JXUM01035048 JXUM01035049 KQ561046 KXJ79901.1 GFDL01008088 JAV26957.1 DS231813 EDS25757.1 GL442284 EFN63667.1 CH477433 EAT41005.1 KQ414657 KOC65831.1 GL451577 EFN78977.1 AAZX01007073 AAZX01015686 AAZX01017394 AXCN02000966 KQ981285 KYN43217.1 CVRI01000059 CRL03032.1 KM507105 AKM28429.1 KQ978344 KYM94805.1 UFQT01001933 SSX32172.1 AAAB01008882 EAA08723.4 GGFL01001029 MBW65207.1 GGFL01001030 MBW65208.1 ATLV01023054 KE525340 KFB48359.1 ADTU01002785 ADTU01002786 ADTU01002787 ADTU01002788 ADTU01002789 GGFK01010064 MBW43385.1 GBHO01017401 GDHC01017279 GDHC01000397 JAG26203.1 JAQ01350.1 JAQ18232.1 GGFM01004431 MBW25182.1 KQ976693 KYM77648.1 KQ982431 KYQ56455.1 ABLF02025451 GBRD01017873 JAG47954.1 GL888065 EGI68412.1 DS235442 EEB15795.1 KK107231 QOIP01000009 EZA55072.1 RLU18933.1 KK853284 KDR08980.1 GFXV01006855 MBW18660.1 ACPB03006494 GDAI01000228 JAI17375.1 GANO01001369 JAB58502.1 GDHC01018237 JAQ00392.1 GFJQ02001067 JAW05903.1 GBHO01017402 GBHO01017399 JAG26202.1 JAG26205.1 CP012524 ALC41817.1 NEVH01006994 PNF36111.1 GBRD01017872 JAG47955.1 CH933808 EDW08111.2 GECL01002156 JAP03968.1 GEDC01029613 GEDC01020895 JAS07685.1 JAS16403.1 GFTR01004661 JAW11765.1 CH940662 EDW58718.1 CRL03835.1 CH916367 EDW01213.1 GEBQ01026846 JAT13131.1 KRF78429.1 KK119043 KFM74477.1 GAMC01005437 GAMC01005435 JAC01121.1 GECZ01006414 JAS63355.1 KRG03912.1 GEDC01028997 GEDC01004598 JAS08301.1 JAS32700.1 GFWV01022020 MAA46748.1 EZ422699 ADD18975.1 GBXI01000199 JAD14093.1 GAMC01005436 JAC01120.1 OUUW01000002 SPP76338.1 GEDC01021603 GEDC01011468 JAS15695.1 JAS25830.1 KPJ10451.1 GFDG01004345 JAV14454.1 PZQS01000003 PVD33358.1 CH963849 EDW74093.1 GFDG01000241 JAV18558.1 GFDG01000242 JAV18557.1 GFDG01000240 JAV18559.1 CH902619 KPU76357.1 GL732548 EFX80182.1
AK401932 BAM18554.1 RSAL01000136 RVE46219.1 GDQN01002683 GDQN01001775 JAT88371.1 JAT89279.1 NWSH01003423 PCG66334.1 AGBW02005277 OWR55305.1 GAIX01005398 JAA87162.1 ODYU01000021 SOQ34090.1 OWR55302.1 RVE46217.1 NWSH01000947 PCG73418.1 NNAY01000614 OXU27387.1 JXUM01023906 JXUM01023907 JXUM01023908 JXUM01023909 GAPW01002201 KQ560674 JAC11397.1 KXJ81330.1 JXUM01035044 JXUM01035045 JXUM01035046 JXUM01035047 JXUM01035048 JXUM01035049 KQ561046 KXJ79901.1 GFDL01008088 JAV26957.1 DS231813 EDS25757.1 GL442284 EFN63667.1 CH477433 EAT41005.1 KQ414657 KOC65831.1 GL451577 EFN78977.1 AAZX01007073 AAZX01015686 AAZX01017394 AXCN02000966 KQ981285 KYN43217.1 CVRI01000059 CRL03032.1 KM507105 AKM28429.1 KQ978344 KYM94805.1 UFQT01001933 SSX32172.1 AAAB01008882 EAA08723.4 GGFL01001029 MBW65207.1 GGFL01001030 MBW65208.1 ATLV01023054 KE525340 KFB48359.1 ADTU01002785 ADTU01002786 ADTU01002787 ADTU01002788 ADTU01002789 GGFK01010064 MBW43385.1 GBHO01017401 GDHC01017279 GDHC01000397 JAG26203.1 JAQ01350.1 JAQ18232.1 GGFM01004431 MBW25182.1 KQ976693 KYM77648.1 KQ982431 KYQ56455.1 ABLF02025451 GBRD01017873 JAG47954.1 GL888065 EGI68412.1 DS235442 EEB15795.1 KK107231 QOIP01000009 EZA55072.1 RLU18933.1 KK853284 KDR08980.1 GFXV01006855 MBW18660.1 ACPB03006494 GDAI01000228 JAI17375.1 GANO01001369 JAB58502.1 GDHC01018237 JAQ00392.1 GFJQ02001067 JAW05903.1 GBHO01017402 GBHO01017399 JAG26202.1 JAG26205.1 CP012524 ALC41817.1 NEVH01006994 PNF36111.1 GBRD01017872 JAG47955.1 CH933808 EDW08111.2 GECL01002156 JAP03968.1 GEDC01029613 GEDC01020895 JAS07685.1 JAS16403.1 GFTR01004661 JAW11765.1 CH940662 EDW58718.1 CRL03835.1 CH916367 EDW01213.1 GEBQ01026846 JAT13131.1 KRF78429.1 KK119043 KFM74477.1 GAMC01005437 GAMC01005435 JAC01121.1 GECZ01006414 JAS63355.1 KRG03912.1 GEDC01028997 GEDC01004598 JAS08301.1 JAS32700.1 GFWV01022020 MAA46748.1 EZ422699 ADD18975.1 GBXI01000199 JAD14093.1 GAMC01005436 JAC01120.1 OUUW01000002 SPP76338.1 GEDC01021603 GEDC01011468 JAS15695.1 JAS25830.1 KPJ10451.1 GFDG01004345 JAV14454.1 PZQS01000003 PVD33358.1 CH963849 EDW74093.1 GFDG01000241 JAV18558.1 GFDG01000242 JAV18557.1 GFDG01000240 JAV18559.1 CH902619 KPU76357.1 GL732548 EFX80182.1
Proteomes
UP000053268
UP000053240
UP000283053
UP000218220
UP000007151
UP000215335
+ More
UP000069940 UP000249989 UP000002320 UP000000311 UP000008820 UP000053825 UP000008237 UP000076408 UP000002358 UP000075886 UP000078541 UP000075903 UP000183832 UP000069272 UP000075900 UP000078542 UP000007062 UP000030765 UP000075880 UP000005205 UP000075920 UP000078540 UP000075809 UP000007819 UP000007755 UP000009046 UP000053097 UP000279307 UP000027135 UP000015103 UP000092553 UP000235965 UP000009192 UP000008792 UP000079169 UP000001070 UP000075884 UP000054359 UP000075885 UP000192221 UP000268350 UP000092445 UP000245119 UP000007798 UP000007801 UP000000305
UP000069940 UP000249989 UP000002320 UP000000311 UP000008820 UP000053825 UP000008237 UP000076408 UP000002358 UP000075886 UP000078541 UP000075903 UP000183832 UP000069272 UP000075900 UP000078542 UP000007062 UP000030765 UP000075880 UP000005205 UP000075920 UP000078540 UP000075809 UP000007819 UP000007755 UP000009046 UP000053097 UP000279307 UP000027135 UP000015103 UP000092553 UP000235965 UP000009192 UP000008792 UP000079169 UP000001070 UP000075884 UP000054359 UP000075885 UP000192221 UP000268350 UP000092445 UP000245119 UP000007798 UP000007801 UP000000305
Pfam
PF01569 PAP2
SUPFAM
SSF48317
SSF48317
ProteinModelPortal
A0A194QID4
A0A194QYA8
A0A2H1V758
I4DKW4
A0A3S2LGJ6
A0A1E1WNF3
+ More
A0A2A4J496 A0A212FNJ1 S4PXA1 A0A2H1UZR4 A0A212FNH6 A0A3S2LX18 A0A2A4JNX9 A0A232FAM8 A0A023EPH9 A0A182HE45 A0A1Q3FHC6 B0VZ03 E2ASD3 Q172P3 A0A0L7R4Y5 E2BZ71 A0A182YQY6 K7J4V2 A0A182QVZ1 A0A195FSE8 A0A182VKV9 A0A1J1ISA8 A0A182FLB6 A0A182RUT8 A0A0G3YL55 A0A195C2I9 A0A336MP79 Q7QAY1 A0A2M4CIP2 A0A2M4CJY6 A0A084WDR5 A0A182JFU0 A0A158NNZ1 A0A182WM77 A0A2M4ARF8 A0A0A9Y520 A0A2M3Z9K7 A0A151HZK1 A0A151X7U8 J9JYU5 A0A0K8S456 F4WBK7 E0VQY9 A0A026WG83 A0A067QTP7 A0A2H8TXM7 T1HKB2 A0A0K8TTU9 U5EWL0 A0A146KZH3 A0A1Z5LES4 A0A0A9Y9L2 A0A0M4EKC2 A0A2J7R5L3 A0A0K8S5D9 B4KQ09 A0A0V0G785 A0A1B6C395 A0A224XH03 B4MDV3 A0A3Q0IQY3 A0A1J1IU97 B4J876 A0A182NLS8 A0A1B6KNY2 A0A0Q9W0A7 A0A087UAT8 W8BIL3 A0A1B6GLP9 A0A0Q9XJA5 A0A1B6E443 A0A182PVZ8 A0A1W4V011 A0A293N3I0 D3TMP7 A0A0A1XTI2 W8CAT4 A0A3B0JYZ9 A0A1B6CQX8 A0A194R3N3 A0A1L8E725 A0A1B0AHS6 A0A2T7PIV9 B4MPQ4 A0A1L8EIS0 A0A1L8EIM8 A0A1L8EIW2 A0A0P8XP46 E9GJV9
A0A2A4J496 A0A212FNJ1 S4PXA1 A0A2H1UZR4 A0A212FNH6 A0A3S2LX18 A0A2A4JNX9 A0A232FAM8 A0A023EPH9 A0A182HE45 A0A1Q3FHC6 B0VZ03 E2ASD3 Q172P3 A0A0L7R4Y5 E2BZ71 A0A182YQY6 K7J4V2 A0A182QVZ1 A0A195FSE8 A0A182VKV9 A0A1J1ISA8 A0A182FLB6 A0A182RUT8 A0A0G3YL55 A0A195C2I9 A0A336MP79 Q7QAY1 A0A2M4CIP2 A0A2M4CJY6 A0A084WDR5 A0A182JFU0 A0A158NNZ1 A0A182WM77 A0A2M4ARF8 A0A0A9Y520 A0A2M3Z9K7 A0A151HZK1 A0A151X7U8 J9JYU5 A0A0K8S456 F4WBK7 E0VQY9 A0A026WG83 A0A067QTP7 A0A2H8TXM7 T1HKB2 A0A0K8TTU9 U5EWL0 A0A146KZH3 A0A1Z5LES4 A0A0A9Y9L2 A0A0M4EKC2 A0A2J7R5L3 A0A0K8S5D9 B4KQ09 A0A0V0G785 A0A1B6C395 A0A224XH03 B4MDV3 A0A3Q0IQY3 A0A1J1IU97 B4J876 A0A182NLS8 A0A1B6KNY2 A0A0Q9W0A7 A0A087UAT8 W8BIL3 A0A1B6GLP9 A0A0Q9XJA5 A0A1B6E443 A0A182PVZ8 A0A1W4V011 A0A293N3I0 D3TMP7 A0A0A1XTI2 W8CAT4 A0A3B0JYZ9 A0A1B6CQX8 A0A194R3N3 A0A1L8E725 A0A1B0AHS6 A0A2T7PIV9 B4MPQ4 A0A1L8EIS0 A0A1L8EIM8 A0A1L8EIW2 A0A0P8XP46 E9GJV9
Ontologies
PATHWAY
00561
Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00564 Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00600 Sphingolipid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00564 Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00600 Sphingolipid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
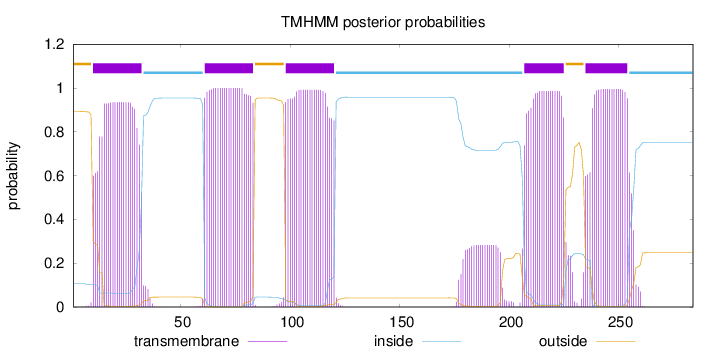
Topology
Length:
284
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
110.05909
Exp number, first 60 AAs:
19.83205
Total prob of N-in:
0.10624
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 60
TMhelix
61 - 83
outside
84 - 97
TMhelix
98 - 120
inside
121 - 206
TMhelix
207 - 225
outside
226 - 234
TMhelix
235 - 254
inside
255 - 284
Population Genetic Test Statistics
Pi
245.263793
Theta
153.664894
Tajima's D
1.527077
CLR
0.266432
CSRT
0.791410429478526
Interpretation
Uncertain
