Gene
KWMTBOMO11692 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004140
Annotation
PREDICTED:_probable_methylcrotonoyl-CoA_carboxylase_beta_chain?_mitochondrial_[Amyelois_transitella]
Full name
Probable methylcrotonoyl-CoA carboxylase beta chain, mitochondrial
Alternative Name
3-methylcrotonyl-CoA carboxylase 2
3-methylcrotonyl-CoA carboxylase non-biotin-containing subunit
3-methylcrotonyl-CoA:carbon dioxide ligase subunit beta
3-methylcrotonyl-CoA carboxylase non-biotin-containing subunit
3-methylcrotonyl-CoA:carbon dioxide ligase subunit beta
Location in the cell
Cytoplasmic Reliability : 1.513 Mitochondrial Reliability : 1.637 Peroxisomal Reliability : 1.075
Sequence
CDS
ATGTTTAAACTCGTCAGAAGCATACGCCGACTGAGCGCACAATGCCGGCACCACAGCCACGCGACCGTCATCGGTACTGAGCCGAACCGAAGTGATCCTTACTTTCAAGAGAATAAGGCGAAAATGGATGAACTGATCGACGAGCTTCAACAGAAAACCGGTGACGCTATCAGAGGTGGGCCAGAGGACGCAGTAAAGAGGCACACGGCGAGAGGAAAATTGCTTGTCAAAGACAGGATTAACCGGTTAGTGGACGAGGGCTCCGATGTATTGGAATTGAGCGCGCTCGCCGCCGCCGACATGTACAAGGGCCAGGTTCCGAGCGCCGGTATCGTCACCGCTATCGGCAAAGTTCACGGTCGCGATTGCATGATCGTCGCTAATGATGCAACCGTTAAAGGGGGAACTTATTTTCCGATTACCGTGAAGAAGCACTTACGCGCCCAGTCGATCGCACAGGAATGTCGGCTTCCGTGCATTTATCTTGTTGATTCTGGTGGAGCCCATCTCCCTGATCAGGCTGATGTGTTTCCGGACCGCGAACACTTCGGAAGAATATTTTACAATCAAGCTAATATGTCAGCTGAGGGTATACCGCAGATATCTGTTGTCATGGGATCGTGCACTGCGGGAGGAGCGTACATCCCTAGTATGTCAGACGAGAGTATTATAATAAAAAATCAAGGAACCATTTTCTTGGCCGGCCCTCCATTAGTCAAAGCCGCTACTGGTGAATCAGTTTCTGCTGAAGATTTAGGTGGAGCTGACCTACATTGCCGGCAATCAGGAGTAACGGATCATTATGCTCTAGATGATGAGCATGCTTTACATTTAGCTAGGAATGTCGTGGCCAACCTCAATTGGAACAATGACCAAAGAGCTAGAGTTTATTCCTCGAAAATTGAGGAGCCTGTTCATGACATTCAGGATTTACACGGAATAGTAGGTGCTAATATTCAGAGACCTTTCGACATTAGAGAGGTTATATCCCGGATAGTTGACGGAAGTAGATTTCATGAATTCAAGCAACTTTATGGGGACACTTTAGTCTGCGGCTTCGCCACATTGTATGGACACCCAGTAGGTGTAATCGGGAACAACGGCGTTCTTATGTCCGAATCAGCTTTGAAGGGATCCCATTTCATTCAGCTGTGCGCTGCTAGGAAGATTCCTTTAGTGTTCCTGCAAAACATTACAGGTTTTATGGTAGGAAAAGACGCTGAAGCTGGCGGTATTGCTAAAAATGGAGCTAAAATGGTAACTGCGGTCAGTTGTTTCAAAGGACCGAAAATAACTGTGATTGTTGGGGGAAGTTTCGGTGCTGGGAACTATGGAATGTGTGGGCGAGCGTATTCGCCGAGTTTTCTTTTCATGTGGCCCAACGCAAGAATATCCGTGATGGGGGGACCACAAGCTGCAACAGTTCTCTCCTTGGTCGCCAAAGAAAAGGCTCAGCGTGAAGGGAAACCGTGGACTGATGAAGATGAAAAGAAAGTCAGGGGTCCATTAGAAGCGCAATTTGAGAAAGAGGGTCTTCCTTACTACAGTACAGCAAGGCTGTGGGATGATGGCATTGTAGCTCCGAAAGATACGAGAAAAATTATAGGCCTAAGTTTATCGGCAGCTCTTAACGCACCGTTTAGAGAAAGCAAATTCGGCATCTTTAGAATGTGA
Protein
MFKLVRSIRRLSAQCRHHSHATVIGTEPNRSDPYFQENKAKMDELIDELQQKTGDAIRGGPEDAVKRHTARGKLLVKDRINRLVDEGSDVLELSALAAADMYKGQVPSAGIVTAIGKVHGRDCMIVANDATVKGGTYFPITVKKHLRAQSIAQECRLPCIYLVDSGGAHLPDQADVFPDREHFGRIFYNQANMSAEGIPQISVVMGSCTAGGAYIPSMSDESIIIKNQGTIFLAGPPLVKAATGESVSAEDLGGADLHCRQSGVTDHYALDDEHALHLARNVVANLNWNNDQRARVYSSKIEEPVHDIQDLHGIVGANIQRPFDIREVISRIVDGSRFHEFKQLYGDTLVCGFATLYGHPVGVIGNNGVLMSESALKGSHFIQLCAARKIPLVFLQNITGFMVGKDAEAGGIAKNGAKMVTAVSCFKGPKITVIVGGSFGAGNYGMCGRAYSPSFLFMWPNARISVMGGPQAATVLSLVAKEKAQREGKPWTDEDEKKVRGPLEAQFEKEGLPYYSTARLWDDGIVAPKDTRKIIGLSLSAALNAPFRESKFGIFRM
Summary
Description
Carboxyltransferase subunit of the 3-methylcrotonyl-CoA carboxylase, an enzyme that catalyzes the conversion of 3-methylcrotonyl-CoA to 3-methylglutaconyl-CoA, a critical step for leucine and isovaleric acid catabolism (By similarity). Vital for adult survival.
Catalytic Activity
3-methylbut-2-enoyl-CoA + ATP + hydrogencarbonate = ADP + H(+) + phosphate + trans-3-methylglutaconyl-CoA
Similarity
Belongs to the bacterial ribosomal protein bL20 family.
Belongs to the AccD/PCCB family.
Belongs to the AccD/PCCB family.
Keywords
ATP-binding
Complete proteome
Ligase
Mitochondrion
Nucleotide-binding
Reference proteome
Transit peptide
Feature
chain Probable methylcrotonoyl-CoA carboxylase beta chain, mitochondrial
Uniprot
H9J3Q3
A0A2A4JFV6
A0A2H1UZQ6
A0A194QCR8
A0A194QYT3
A0A0L7LN29
+ More
A0A212FNK2 A0A182IU36 B0WSS2 A0A1Q3EXM2 A0A084VTA7 Q16HN6 A0A1S4G0N4 A0A182G411 A0A182RSY4 A0A2M4AF07 W5J6M0 A0A2M4AF18 A0A182GIX5 A0A182LV31 A0A182VZQ0 A0A182TRV2 A0A023F596 A0A182Y3W0 Q7QCQ8 A0A1I8JSV1 A0A182XIU1 A0A023EU43 A0A182LBZ0 A0A2M3Z8F4 A0A182UXG3 A0A2M4BI51 A0A224XE59 A0A182JW06 A0A1W4V2F0 A0A1B0CDN4 A0A182QJQ2 A0A2M4BI55 B4KNH7 A0A034WCJ5 H0RNI1 A0A034WG38 B4HQB5 B4QDC4 Q9V9A7 A0A182P505 A0A0V0GAJ7 T1IDW8 B3MIN7 A0A182NQG6 A0A0P4VNG7 T1PBE2 B3NB25 A0A0K8UDW6 A0A0L0BMS4 B4P109 A0A0A1XIC4 C3YKH0 B4MY47 A0A182TA99 A0A0M4EL13 A0A1L8E2U2 A0A2L2YBJ1 H2ZJ79 A0A131Y2P7 R7V6W0 A0A1S3J4A6 G3MGR3 A0A0P4VYB1 H2ZJ78 A0A0K8RLB7 A0A1L8E2B7 A0A026W3Y9 W8CE36 A0A023GNV0 A0A2J7R0N0 A0A1E1XFD9 B4GC88 A0A1I8P2J9 A0A0K2TCS0 B4LK16 A0A1A9V4S7 E2A1E4 Q292Y6 A0A336M791 D3TNT3 A0A1B0EUD2 U5ESP2 A0A067RIX1 A0A1A9ZX14 A0A1W4VFL1 A0A3B0J218 A0A1A9WNW0 A0A2R5LHB1 A0A1Z5LCR5 F6T882 A0A2I4CWA8 A0A226CX02 E0VID1 A0A1S3J3R7
A0A212FNK2 A0A182IU36 B0WSS2 A0A1Q3EXM2 A0A084VTA7 Q16HN6 A0A1S4G0N4 A0A182G411 A0A182RSY4 A0A2M4AF07 W5J6M0 A0A2M4AF18 A0A182GIX5 A0A182LV31 A0A182VZQ0 A0A182TRV2 A0A023F596 A0A182Y3W0 Q7QCQ8 A0A1I8JSV1 A0A182XIU1 A0A023EU43 A0A182LBZ0 A0A2M3Z8F4 A0A182UXG3 A0A2M4BI51 A0A224XE59 A0A182JW06 A0A1W4V2F0 A0A1B0CDN4 A0A182QJQ2 A0A2M4BI55 B4KNH7 A0A034WCJ5 H0RNI1 A0A034WG38 B4HQB5 B4QDC4 Q9V9A7 A0A182P505 A0A0V0GAJ7 T1IDW8 B3MIN7 A0A182NQG6 A0A0P4VNG7 T1PBE2 B3NB25 A0A0K8UDW6 A0A0L0BMS4 B4P109 A0A0A1XIC4 C3YKH0 B4MY47 A0A182TA99 A0A0M4EL13 A0A1L8E2U2 A0A2L2YBJ1 H2ZJ79 A0A131Y2P7 R7V6W0 A0A1S3J4A6 G3MGR3 A0A0P4VYB1 H2ZJ78 A0A0K8RLB7 A0A1L8E2B7 A0A026W3Y9 W8CE36 A0A023GNV0 A0A2J7R0N0 A0A1E1XFD9 B4GC88 A0A1I8P2J9 A0A0K2TCS0 B4LK16 A0A1A9V4S7 E2A1E4 Q292Y6 A0A336M791 D3TNT3 A0A1B0EUD2 U5ESP2 A0A067RIX1 A0A1A9ZX14 A0A1W4VFL1 A0A3B0J218 A0A1A9WNW0 A0A2R5LHB1 A0A1Z5LCR5 F6T882 A0A2I4CWA8 A0A226CX02 E0VID1 A0A1S3J3R7
EC Number
6.4.1.4
Pubmed
19121390
26354079
26227816
22118469
24438588
17510324
+ More
26483478 20920257 23761445 25474469 25244985 12364791 14747013 17210077 24945155 20966253 17994087 25348373 22936249 10731132 12537572 12537569 9584098 27129103 25315136 26108605 17550304 25830018 18563158 26561354 23254933 22216098 24508170 30249741 24495485 28503490 20798317 15632085 20353571 24845553 28528879 17495919 20566863
26483478 20920257 23761445 25474469 25244985 12364791 14747013 17210077 24945155 20966253 17994087 25348373 22936249 10731132 12537572 12537569 9584098 27129103 25315136 26108605 17550304 25830018 18563158 26561354 23254933 22216098 24508170 30249741 24495485 28503490 20798317 15632085 20353571 24845553 28528879 17495919 20566863
EMBL
BABH01021743
NWSH01001673
PCG70454.1
ODYU01000021
SOQ34085.1
KQ459185
+ More
KPJ03214.1 KQ460949 KPJ10454.1 JTDY01000575 KOB76616.1 AGBW02005277 OWR55307.1 DS232075 EDS34031.1 GFDL01014986 JAV20059.1 ATLV01016287 KE525074 KFB41201.1 CH478156 EAT33766.1 JXUM01041704 JXUM01041705 KQ561294 KXJ79084.1 GGFK01006045 MBW39366.1 ADMH02002131 ETN58500.1 GGFK01006053 MBW39374.1 JXUM01067006 JXUM01067007 JXUM01067008 KQ562428 KXJ75934.1 AXCM01000485 GBBI01002448 JAC16264.1 AAAB01008859 EAA07889.3 GAPW01000700 JAC12898.1 GGFM01004056 MBW24807.1 GGFJ01003598 MBW52739.1 GFTR01007142 JAW09284.1 AJWK01008032 AXCN02001539 GGFJ01003599 MBW52740.1 CH933808 EDW08936.1 GAKP01005676 JAC53276.1 BT132903 AEV66553.1 GAKP01005675 JAC53277.1 CH480816 EDW46654.1 CM000362 CM002911 EDX05911.1 KMY91800.1 AE013599 AF006654 AY084145 AAM70824.1 GECL01001144 JAP04980.1 ACPB03019345 CH902619 EDV38113.1 GDKW01002112 JAI54483.1 KA645455 AFP60084.1 CH954177 EDV59790.1 GDHF01027568 JAI24746.1 JRES01001644 KNC21238.1 CM000157 EDW89083.1 GBXI01004054 JAD10238.1 GG666522 EEN59214.1 CH963894 EDW77036.1 CP012524 ALC42180.1 GFDF01001269 JAV12815.1 IAAA01012927 IAAA01012928 LAA05519.1 GEFM01003018 JAP72778.1 AMQN01000703 KB294417 ELU14608.1 JO841064 AEO32681.1 GDRN01108181 JAI57301.1 GADI01001922 JAA71886.1 GFDF01001270 JAV12814.1 KK107447 QOIP01000001 EZA50718.1 RLU26971.1 GAMC01000881 JAC05675.1 GBBM01000688 JAC34730.1 NEVH01008227 PNF34387.1 GFAC01001201 JAT97987.1 CH479181 EDW31406.1 HACA01005900 CDW23261.1 CH940648 EDW61670.1 GL435760 EFN72770.1 CM000071 EAL24725.1 UFQT01000494 SSX24739.1 EZ423085 ADD19361.1 CCAG010015290 GANO01002266 JAB57605.1 KK852657 KDR19223.1 OUUW01000001 SPP73032.1 GGLE01004743 MBY08869.1 GFJQ02001757 JAW05213.1 LNIX01000060 OXA37290.1 DS235196 EEB13137.1
KPJ03214.1 KQ460949 KPJ10454.1 JTDY01000575 KOB76616.1 AGBW02005277 OWR55307.1 DS232075 EDS34031.1 GFDL01014986 JAV20059.1 ATLV01016287 KE525074 KFB41201.1 CH478156 EAT33766.1 JXUM01041704 JXUM01041705 KQ561294 KXJ79084.1 GGFK01006045 MBW39366.1 ADMH02002131 ETN58500.1 GGFK01006053 MBW39374.1 JXUM01067006 JXUM01067007 JXUM01067008 KQ562428 KXJ75934.1 AXCM01000485 GBBI01002448 JAC16264.1 AAAB01008859 EAA07889.3 GAPW01000700 JAC12898.1 GGFM01004056 MBW24807.1 GGFJ01003598 MBW52739.1 GFTR01007142 JAW09284.1 AJWK01008032 AXCN02001539 GGFJ01003599 MBW52740.1 CH933808 EDW08936.1 GAKP01005676 JAC53276.1 BT132903 AEV66553.1 GAKP01005675 JAC53277.1 CH480816 EDW46654.1 CM000362 CM002911 EDX05911.1 KMY91800.1 AE013599 AF006654 AY084145 AAM70824.1 GECL01001144 JAP04980.1 ACPB03019345 CH902619 EDV38113.1 GDKW01002112 JAI54483.1 KA645455 AFP60084.1 CH954177 EDV59790.1 GDHF01027568 JAI24746.1 JRES01001644 KNC21238.1 CM000157 EDW89083.1 GBXI01004054 JAD10238.1 GG666522 EEN59214.1 CH963894 EDW77036.1 CP012524 ALC42180.1 GFDF01001269 JAV12815.1 IAAA01012927 IAAA01012928 LAA05519.1 GEFM01003018 JAP72778.1 AMQN01000703 KB294417 ELU14608.1 JO841064 AEO32681.1 GDRN01108181 JAI57301.1 GADI01001922 JAA71886.1 GFDF01001270 JAV12814.1 KK107447 QOIP01000001 EZA50718.1 RLU26971.1 GAMC01000881 JAC05675.1 GBBM01000688 JAC34730.1 NEVH01008227 PNF34387.1 GFAC01001201 JAT97987.1 CH479181 EDW31406.1 HACA01005900 CDW23261.1 CH940648 EDW61670.1 GL435760 EFN72770.1 CM000071 EAL24725.1 UFQT01000494 SSX24739.1 EZ423085 ADD19361.1 CCAG010015290 GANO01002266 JAB57605.1 KK852657 KDR19223.1 OUUW01000001 SPP73032.1 GGLE01004743 MBY08869.1 GFJQ02001757 JAW05213.1 LNIX01000060 OXA37290.1 DS235196 EEB13137.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000075880 UP000002320 UP000030765 UP000008820 UP000069940 UP000249989 UP000075900 UP000000673 UP000075883 UP000075920 UP000075902 UP000076408 UP000007062 UP000069272 UP000076407 UP000075882 UP000075903 UP000075881 UP000192221 UP000092461 UP000075886 UP000009192 UP000001292 UP000000304 UP000000803 UP000075885 UP000015103 UP000007801 UP000075884 UP000095301 UP000008711 UP000037069 UP000002282 UP000001554 UP000007798 UP000075901 UP000092553 UP000007875 UP000014760 UP000085678 UP000053097 UP000279307 UP000235965 UP000008744 UP000095300 UP000008792 UP000078200 UP000000311 UP000001819 UP000092444 UP000027135 UP000092445 UP000268350 UP000091820 UP000002280 UP000192220 UP000198287 UP000009046
UP000075880 UP000002320 UP000030765 UP000008820 UP000069940 UP000249989 UP000075900 UP000000673 UP000075883 UP000075920 UP000075902 UP000076408 UP000007062 UP000069272 UP000076407 UP000075882 UP000075903 UP000075881 UP000192221 UP000092461 UP000075886 UP000009192 UP000001292 UP000000304 UP000000803 UP000075885 UP000015103 UP000007801 UP000075884 UP000095301 UP000008711 UP000037069 UP000002282 UP000001554 UP000007798 UP000075901 UP000092553 UP000007875 UP000014760 UP000085678 UP000053097 UP000279307 UP000235965 UP000008744 UP000095300 UP000008792 UP000078200 UP000000311 UP000001819 UP000092444 UP000027135 UP000092445 UP000268350 UP000091820 UP000002280 UP000192220 UP000198287 UP000009046
Interpro
Gene 3D
ProteinModelPortal
H9J3Q3
A0A2A4JFV6
A0A2H1UZQ6
A0A194QCR8
A0A194QYT3
A0A0L7LN29
+ More
A0A212FNK2 A0A182IU36 B0WSS2 A0A1Q3EXM2 A0A084VTA7 Q16HN6 A0A1S4G0N4 A0A182G411 A0A182RSY4 A0A2M4AF07 W5J6M0 A0A2M4AF18 A0A182GIX5 A0A182LV31 A0A182VZQ0 A0A182TRV2 A0A023F596 A0A182Y3W0 Q7QCQ8 A0A1I8JSV1 A0A182XIU1 A0A023EU43 A0A182LBZ0 A0A2M3Z8F4 A0A182UXG3 A0A2M4BI51 A0A224XE59 A0A182JW06 A0A1W4V2F0 A0A1B0CDN4 A0A182QJQ2 A0A2M4BI55 B4KNH7 A0A034WCJ5 H0RNI1 A0A034WG38 B4HQB5 B4QDC4 Q9V9A7 A0A182P505 A0A0V0GAJ7 T1IDW8 B3MIN7 A0A182NQG6 A0A0P4VNG7 T1PBE2 B3NB25 A0A0K8UDW6 A0A0L0BMS4 B4P109 A0A0A1XIC4 C3YKH0 B4MY47 A0A182TA99 A0A0M4EL13 A0A1L8E2U2 A0A2L2YBJ1 H2ZJ79 A0A131Y2P7 R7V6W0 A0A1S3J4A6 G3MGR3 A0A0P4VYB1 H2ZJ78 A0A0K8RLB7 A0A1L8E2B7 A0A026W3Y9 W8CE36 A0A023GNV0 A0A2J7R0N0 A0A1E1XFD9 B4GC88 A0A1I8P2J9 A0A0K2TCS0 B4LK16 A0A1A9V4S7 E2A1E4 Q292Y6 A0A336M791 D3TNT3 A0A1B0EUD2 U5ESP2 A0A067RIX1 A0A1A9ZX14 A0A1W4VFL1 A0A3B0J218 A0A1A9WNW0 A0A2R5LHB1 A0A1Z5LCR5 F6T882 A0A2I4CWA8 A0A226CX02 E0VID1 A0A1S3J3R7
A0A212FNK2 A0A182IU36 B0WSS2 A0A1Q3EXM2 A0A084VTA7 Q16HN6 A0A1S4G0N4 A0A182G411 A0A182RSY4 A0A2M4AF07 W5J6M0 A0A2M4AF18 A0A182GIX5 A0A182LV31 A0A182VZQ0 A0A182TRV2 A0A023F596 A0A182Y3W0 Q7QCQ8 A0A1I8JSV1 A0A182XIU1 A0A023EU43 A0A182LBZ0 A0A2M3Z8F4 A0A182UXG3 A0A2M4BI51 A0A224XE59 A0A182JW06 A0A1W4V2F0 A0A1B0CDN4 A0A182QJQ2 A0A2M4BI55 B4KNH7 A0A034WCJ5 H0RNI1 A0A034WG38 B4HQB5 B4QDC4 Q9V9A7 A0A182P505 A0A0V0GAJ7 T1IDW8 B3MIN7 A0A182NQG6 A0A0P4VNG7 T1PBE2 B3NB25 A0A0K8UDW6 A0A0L0BMS4 B4P109 A0A0A1XIC4 C3YKH0 B4MY47 A0A182TA99 A0A0M4EL13 A0A1L8E2U2 A0A2L2YBJ1 H2ZJ79 A0A131Y2P7 R7V6W0 A0A1S3J4A6 G3MGR3 A0A0P4VYB1 H2ZJ78 A0A0K8RLB7 A0A1L8E2B7 A0A026W3Y9 W8CE36 A0A023GNV0 A0A2J7R0N0 A0A1E1XFD9 B4GC88 A0A1I8P2J9 A0A0K2TCS0 B4LK16 A0A1A9V4S7 E2A1E4 Q292Y6 A0A336M791 D3TNT3 A0A1B0EUD2 U5ESP2 A0A067RIX1 A0A1A9ZX14 A0A1W4VFL1 A0A3B0J218 A0A1A9WNW0 A0A2R5LHB1 A0A1Z5LCR5 F6T882 A0A2I4CWA8 A0A226CX02 E0VID1 A0A1S3J3R7
PDB
3U9T
E-value=0,
Score=1762
Ontologies
PATHWAY
GO
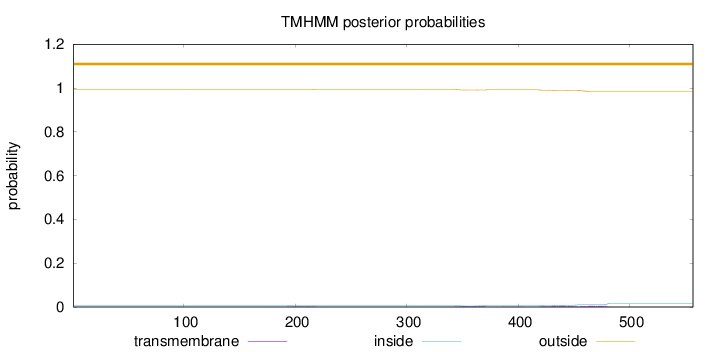
Topology
Subcellular location
Mitochondrion matrix
Length:
557
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.40089
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00803
outside
1 - 557
Population Genetic Test Statistics
Pi
231.050436
Theta
223.008837
Tajima's D
0.219368
CLR
0.013185
CSRT
0.432328383580821
Interpretation
Uncertain
