Gene
KWMTBOMO11690
Pre Gene Modal
BGIBMGA004139
Annotation
PREDICTED:_uncharacterized_protein_LOC101744923_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.978
Sequence
CDS
ATGAACATAAAAGGTTGGAAGAAAACAGAGAACGAAGCGGGATATCCTTGTTACATTAATGAAACTACCAAAAAACAACAATATGATCATCCCGAATTTATTAAATTATGGCAATCACTTTCTGTATACGACGATATAAAATATGCAGTGTACCGTATTGCGTTCAAAATCCATGCTTTGCAAAAGCAAATTCGAGTGCCTCCATTAAGAATCAGTTCTGGAGTTTTTGCTCGGCATCAACTTAGTTTGTCTGAAAGCAGTCTATCATTGGATACTACTGAGCTGGAAGCTGTACTAGCTGATATATATTTTGCTGCAGAAAAAGATGGTCTATTTGATGGGGATGTTGACTTTGCTGTAGATCTCCTCATTAATCTCTTATTAAATGTTTATGATGTTGACCGAAAAGAACCGATACGAGTATTAGCAGCAAAGACCCTTCTTATTGTCCTGAGTAATGATCCAGCATCAGACAAATGGTTTGCACTAGCCAATAGTTGTGCAGATCACAATGGATGCGTGTCTGCCAAGAGATTCTCAGCTTTATTAACGCATATTGTAGCACTGCCGCTATACTGTGGAACATCCTCTGGACAGCTGCAGGATGATGTTCTCTCGTGTTTTGATAAGAATTCGGGTATGTTAGGCGTAAGTTCACGGATGGTAGCCGAATGGGGTGTGCAGTACTGCGCTAGTACAATGTGGCTGTCAGTTTTAGAACGAATCTCAGCTAGTATTGACGATGAGAATACTATTGGTCAATGCGCTGTTTGCTCACAAAACCTAAAGAAGATGCTGAAATTTAAATGTTCAAAATGCAGTGAGGTATATTTTTGTGAGAACTGTTATTTATATGACCGAGGCTTGAATACAGTAAGTGGACACAAGAAGACTCATGTAGTGCAAGAAATTGTTGATGGACAGATTAAATCCTCACAATGCGCGTTCATGAAGAGTATGAGACGATTCTTCTTTTGCACTAAGACTAAGAAGAAGAAGATAGTGAGATCAAAAAAAATCGACCAGTTAGACGGGCCCAGCACTCTAAAACGAAAGAAGGGTAACAAACCAACCATGTTCACTTCAACAGTTGGTAAAGCTTCAGGGAAAGAGGCTGGCAACCCCGCGAACATGCTTCAAGATATTATCATGCAGCTGGAAGCTCAGAATAGAGCTCTTCAAGAATTATCGGTGCAGTTCAAGGACGTTGGAAAGAATGATGATGAACTCAAAACGAAGGTAGATACACATTGGTCACAGATATCTACCCAAATAAATAGACTTAAAGTTTTAAAGGAAAATTTGATTGCACCTCAAAATGTGTCCAACGATACAGACAACAAAGACAAGCCTCAAGCTTTTGACCTGTTCAGTCCCATACCGGTCATCGAAACTCAGACCAAACTGACAGACAAGAACCATCCTAGAGTATTGAGCTTGGACTCCGGGAACTTCTCAGTCGTGTCGAAACAAGAGAGCCAGAACTTGCTGACGTTGTCTGGAGATGCCTTGAAACCGGTGGTTGCTCATGCCACATCAGATAGCATATCCGCTGTTTCTGTGAATGATATCAGCTCGTGGTACAATGAAACGCAGGCTTCCGGCAAGCATGTCCGCCAAGCAAAGGGGCAGCCCCCGGTGGAGGTCCCGCCCCCCGTGGAGGTCCCGCCCCCCTCGGCGGAGGACAGGTTCGCGGCTGACATGCGGCGCGTGGAGCACGGCAATCACAAGATGAAAGAGCTCAATGCGGACCTGGACAAGGTGCTCGACAGGCTGCAGGAAATACTCACACACAACTTCACGATGGACGATTCATGTTTCGATAATAGCCAGCTGCGCGAGACGGCGAATGAAATGGAAGGTCTGTTGGGAACATTGATACGCGGAGTCGAACAGCGAGCGACCCTGGGCAACACTAAGGCGCTCGTGTAA
Protein
MNIKGWKKTENEAGYPCYINETTKKQQYDHPEFIKLWQSLSVYDDIKYAVYRIAFKIHALQKQIRVPPLRISSGVFARHQLSLSESSLSLDTTELEAVLADIYFAAEKDGLFDGDVDFAVDLLINLLLNVYDVDRKEPIRVLAAKTLLIVLSNDPASDKWFALANSCADHNGCVSAKRFSALLTHIVALPLYCGTSSGQLQDDVLSCFDKNSGMLGVSSRMVAEWGVQYCASTMWLSVLERISASIDDENTIGQCAVCSQNLKKMLKFKCSKCSEVYFCENCYLYDRGLNTVSGHKKTHVVQEIVDGQIKSSQCAFMKSMRRFFFCTKTKKKKIVRSKKIDQLDGPSTLKRKKGNKPTMFTSTVGKASGKEAGNPANMLQDIIMQLEAQNRALQELSVQFKDVGKNDDELKTKVDTHWSQISTQINRLKVLKENLIAPQNVSNDTDNKDKPQAFDLFSPIPVIETQTKLTDKNHPRVLSLDSGNFSVVSKQESQNLLTLSGDALKPVVAHATSDSISAVSVNDISSWYNETQASGKHVRQAKGQPPVEVPPPVEVPPPSAEDRFAADMRRVEHGNHKMKELNADLDKVLDRLQEILTHNFTMDDSCFDNSQLRETANEMEGLLGTLIRGVEQRATLGNTKALV
Summary
Uniprot
Pubmed
EMBL
BABH01021741
BABH01021742
ODYU01000021
SOQ34083.1
KQ459185
KPJ03215.1
+ More
AGBW02005277 OWR55309.1 NWSH01001673 PCG70451.1 ODYU01007466 SOQ50267.1 JTDY01000575 KOB76617.1 GEZM01093588 JAV56234.1 KQ971354 EFA06845.2 NEVH01006994 PNF36110.1 GECU01023748 JAS83958.1 KQ976662 KYM78436.1 KQ981864 KYN34358.1 KQ979568 KYN20781.1 KQ982314 KYQ58014.1 KQ435119 KZC14713.1 GL442545 EFN63343.1 GL888818 EGI57801.1 ADTU01019320 KZ288310 PBC28600.1 KQ414583 KOC70737.1 GEDC01028928 GEDC01028619 GEDC01023900 GEDC01019589 JAS08370.1 JAS08679.1 JAS13398.1 JAS17709.1 NNAY01003499 OXU19320.1 KQ977647 KYN00848.1
AGBW02005277 OWR55309.1 NWSH01001673 PCG70451.1 ODYU01007466 SOQ50267.1 JTDY01000575 KOB76617.1 GEZM01093588 JAV56234.1 KQ971354 EFA06845.2 NEVH01006994 PNF36110.1 GECU01023748 JAS83958.1 KQ976662 KYM78436.1 KQ981864 KYN34358.1 KQ979568 KYN20781.1 KQ982314 KYQ58014.1 KQ435119 KZC14713.1 GL442545 EFN63343.1 GL888818 EGI57801.1 ADTU01019320 KZ288310 PBC28600.1 KQ414583 KOC70737.1 GEDC01028928 GEDC01028619 GEDC01023900 GEDC01019589 JAS08370.1 JAS08679.1 JAS13398.1 JAS17709.1 NNAY01003499 OXU19320.1 KQ977647 KYN00848.1
Proteomes
PRIDE
Interpro
CDD
ProteinModelPortal
PDB
1EG4
E-value=3.38285e-21,
Score=253
Ontologies
GO
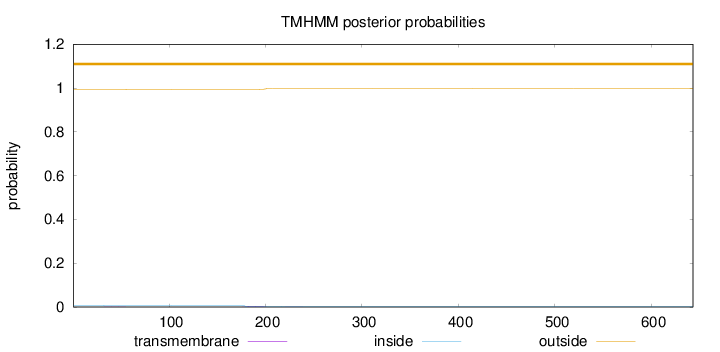
Topology
Length:
643
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08922
Exp number, first 60 AAs:
0.00345
Total prob of N-in:
0.00614
outside
1 - 643
Population Genetic Test Statistics
Pi
192.777561
Theta
169.67725
Tajima's D
0.296929
CLR
0.005667
CSRT
0.449927503624819
Interpretation
Uncertain
