Gene
KWMTBOMO11689 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003972
Annotation
selenophosphate_synthetase_1_[Bombyx_mori]
Full name
Selenide, water dikinase
+ More
GTP:AMP phosphotransferase, mitochondrial
GTP:AMP phosphotransferase, mitochondrial
Alternative Name
Protein patufet
Selenium donor protein
Selenophosphate synthase
dSelD
Adenylate kinase 3
Selenium donor protein
Selenophosphate synthase
dSelD
Adenylate kinase 3
Location in the cell
Cytoplasmic Reliability : 2.514
Sequence
CDS
ATGTCTTATCAAACGAGTGTAGCGCAGGACTCTTTAGCTGCAGCTCAATTAGAAATGGCCGGTAACCCTAATTCAATAGCTTTACGTCGTCCGTTTGACCCGGTGGCGCACGATTTGGAGGCAAGTTTTCGGCTGACACGATTTGCTGATCTAAAGGGAAGAAGTTGCAAAGTACCTCAAGATGTTCTCGGAAAACTTGTTGAGTCTTTACAGCAAGACTATTCGCAAGCGGATCAAGACCAATTTATGCATGTGGCTATCCCGCGCATCGGCATCGGCTTGGACTGCTCGGTGACACCACTTAGACACGGTGGGCTCTGTTTAGTTCAAACTACTGACTTTTTTTACCCATTAGTGGATGACCCATATATGATGGGTAAGATTGCATGTGCCAATGTTCTTAGCGACCTATACGCGATGGGTGTTACGGAGTGCGACAATATGCTCATGTTGTTGGGTGTCTCAACGAAAATGACTGAAAAGGAACGCGACGTCGTTATTCCTCTTATAATGCGCGGGTTCAAAGATTCCGCTTTGGAGGCAGGAACGTCTGTCACTGGTGGGCAAACTGTCATCAACCCCTGGTGCACAATTGGTGGGGTTGCTACCACTATCTGCCAGCCTAATGAGTACATTGTACCTGATAATGCAGTGATGGGTGATGTATTGGTCTTGACAAAACCTCTTGGTACACAAGTTGCAGTAAATGCTCACCAATGGTTGGACCAACCTGAACGCTGGAACAGAATCAAATTAGTTGTATCTGAAGAAGACGTTCGTAAAGCATATCACAGAGCAATGGACTCTATGAGCAGACTCAATCGCATTGCAGCAAGATTGATGCACAAGTACAATGCTCACGGCTCAACAGATGTAACTGGTTTTGGTTTACTGGGTCATGCACAGAACTTGGCATCTCATCAAAAGAATGAAGTATCATTTGTAATCCATAATCTGCCTGTAATTGCCAAGATGGCAGCAGTAGCGAAAGCTTGTGGGAACATGTTTCAATTGCTACAAGGACACGCTCCAGAGACATCTGGTGGACTTCTAATCTGTCTGCCACGAGAACAGGCTGCTGCGTACTGTAAAGATATTGAGAAACAAGAAGGATATCAGGCTTGGATCATTGGTATTGTGGAGAAGGGTAACCGCACAGCCCGCATTATTGATAAACCTCGAGTCATTGAAGTGCCTGCTAAAGATTAA
Protein
MSYQTSVAQDSLAAAQLEMAGNPNSIALRRPFDPVAHDLEASFRLTRFADLKGRSCKVPQDVLGKLVESLQQDYSQADQDQFMHVAIPRIGIGLDCSVTPLRHGGLCLVQTTDFFYPLVDDPYMMGKIACANVLSDLYAMGVTECDNMLMLLGVSTKMTEKERDVVIPLIMRGFKDSALEAGTSVTGGQTVINPWCTIGGVATTICQPNEYIVPDNAVMGDVLVLTKPLGTQVAVNAHQWLDQPERWNRIKLVVSEEDVRKAYHRAMDSMSRLNRIAARLMHKYNAHGSTDVTGFGLLGHAQNLASHQKNEVSFVIHNLPVIAKMAAVAKACGNMFQLLQGHAPETSGGLLICLPREQAAAYCKDIEKQEGYQAWIIGIVEKGNRTARIIDKPRVIEVPAKD
Summary
Description
Synthesizes selenophosphate from selenide and ATP. Essential for progression of the cell cycle by controlling the synthesis of selenoproteins. Plays a role in apoptosis and consequently a role in imaginal disk patterning and growth.
Involved in maintaining the homeostasis of cellular nucleotides by catalyzing the interconversion of nucleoside phosphates. Has GTP:AMP phosphotransferase and ITP:AMP phosphotransferase activities.
Involved in maintaining the homeostasis of cellular nucleotides by catalyzing the interconversion of nucleoside phosphates. Has GTP:AMP phosphotransferase and ITP:AMP phosphotransferase activities.
Catalytic Activity
ATP + H2O + hydrogenselenide = AMP + 2 H(+) + phosphate + selenophosphate
a ribonucleoside 5'-triphosphate + AMP = a ribonucleoside 5'-diphosphate + ADP
a ribonucleoside 5'-triphosphate + AMP = a ribonucleoside 5'-diphosphate + ADP
Subunit
Monomer.
Similarity
Belongs to the selenophosphate synthase 1 family. Class II subfamily.
Belongs to the adenylate kinase family. AK3 subfamily.
Belongs to the adenylate kinase family. AK3 subfamily.
Keywords
ATP-binding
Cell cycle
Complete proteome
Developmental protein
Kinase
Nucleotide-binding
Reference proteome
Selenium
Transferase
Feature
chain Selenide, water dikinase
Uniprot
H9J385
Q3HR35
A0A212FNP3
S4PIS4
A0A194QYB3
A0A194QID8
+ More
F5CBM5 A0A0L7LN24 A0A2H1V151 C4NFK8 D6WQY1 A0A1W4XPA8 J3JZ15 A0A1Y1M7B0 A0A0K8TM22 A0A2J7RSV7 A0A1L8DM15 A0A1A9XPQ8 A0A1B0AM41 A0A1A9WIA8 A0A1A9V7P8 A0A2J7RDB1 A0A1A9ZPC6 D3TNX4 A0A0K8VRH3 A0A034WST6 A0A336KLL5 A0A2P8YTY2 A0A1I8PVW5 A0A0A1WYS7 A0A2P8ZH28 A0A0L0CB70 T1P7N8 A0A1B6FWM1 W8C155 A0A158NS94 B3MDY9 A0A1W4WBB6 B4P736 B3NR64 B4HRA6 B4QFF3 O18373 B4J758 B4LMH5 B4KSK6 B4H519 Q28X55 B4MJM5 A0A3Q0JDN0 A0A3B0JUA7 A0A023EQC1 A0A1J1HJS2 A0A1S3DEJ9 U5EUU9 Q16XK1 B0W2W4 A0A1Q3F0R0 A0A182IXD8 A0A182RJB0 A0A182QEQ1 A0A182W7I0 A0A182M2G5 A0A182XZU5 A0A182NTJ7 A0A182T8H0 Q7Q3N9 A0A1S4GWU2 A0A182USB2 A0A182JWN4 A0A182TUC4 A0A182WT47 A0A182L1Z5 A0A182HV42 A0A182PD02 A0A084WHB0 A0A182F301 R4FNR1 A0A2M4BQD9 W5JSF0 A0A2M4AT88 A0A1B6DYK4 A0A2S2PEK6 A0A2H8TFN3 A0A2S2Q2W3 A0A2A3ELU8 J9K9H8 A0A1B6CFT7 F5CBP8 E0VCB3 A0A146LHN0 A0A0K8S9W8 A0A0A9ZA82 E9ICW7 A0A3L8DKU4 A0A195CX57 A0A195FV21 A0A0P6JZ99 A0A0C9R999 A0A151J815
F5CBM5 A0A0L7LN24 A0A2H1V151 C4NFK8 D6WQY1 A0A1W4XPA8 J3JZ15 A0A1Y1M7B0 A0A0K8TM22 A0A2J7RSV7 A0A1L8DM15 A0A1A9XPQ8 A0A1B0AM41 A0A1A9WIA8 A0A1A9V7P8 A0A2J7RDB1 A0A1A9ZPC6 D3TNX4 A0A0K8VRH3 A0A034WST6 A0A336KLL5 A0A2P8YTY2 A0A1I8PVW5 A0A0A1WYS7 A0A2P8ZH28 A0A0L0CB70 T1P7N8 A0A1B6FWM1 W8C155 A0A158NS94 B3MDY9 A0A1W4WBB6 B4P736 B3NR64 B4HRA6 B4QFF3 O18373 B4J758 B4LMH5 B4KSK6 B4H519 Q28X55 B4MJM5 A0A3Q0JDN0 A0A3B0JUA7 A0A023EQC1 A0A1J1HJS2 A0A1S3DEJ9 U5EUU9 Q16XK1 B0W2W4 A0A1Q3F0R0 A0A182IXD8 A0A182RJB0 A0A182QEQ1 A0A182W7I0 A0A182M2G5 A0A182XZU5 A0A182NTJ7 A0A182T8H0 Q7Q3N9 A0A1S4GWU2 A0A182USB2 A0A182JWN4 A0A182TUC4 A0A182WT47 A0A182L1Z5 A0A182HV42 A0A182PD02 A0A084WHB0 A0A182F301 R4FNR1 A0A2M4BQD9 W5JSF0 A0A2M4AT88 A0A1B6DYK4 A0A2S2PEK6 A0A2H8TFN3 A0A2S2Q2W3 A0A2A3ELU8 J9K9H8 A0A1B6CFT7 F5CBP8 E0VCB3 A0A146LHN0 A0A0K8S9W8 A0A0A9ZA82 E9ICW7 A0A3L8DKU4 A0A195CX57 A0A195FV21 A0A0P6JZ99 A0A0C9R999 A0A151J815
EC Number
2.7.9.3
2.7.4.10
2.7.4.10
Pubmed
19121390
22118469
23622113
26354079
26227816
19442669
+ More
18362917 19820115 22516182 23537049 28004739 26369729 20353571 25348373 29403074 25830018 26108605 25315136 24495485 21347285 17994087 17550304 22936249 9398525 9491069 10731132 12537572 12537569 10444382 11258485 15632085 23185243 24945155 17510324 25244985 12364791 20966253 24438588 20920257 23761445 21664929 20566863 26823975 25401762 21282665 30249741
18362917 19820115 22516182 23537049 28004739 26369729 20353571 25348373 29403074 25830018 26108605 25315136 24495485 21347285 17994087 17550304 22936249 9398525 9491069 10731132 12537572 12537569 10444382 11258485 15632085 23185243 24945155 17510324 25244985 12364791 20966253 24438588 20920257 23761445 21664929 20566863 26823975 25401762 21282665 30249741
EMBL
BABH01021741
DQ202318
ABA43640.1
AGBW02005277
OWR55310.1
GAIX01000089
+ More
JAA92471.1 KQ460949 KPJ10457.1 KQ459185 KPJ03216.1 JF505286 AED87507.1 JTDY01000575 KOB76611.1 ODYU01000021 SOQ34082.1 FJ550361 NWSH01000825 ACR07790.1 PCG73997.1 KQ971351 EFA06522.1 APGK01055449 BT128496 KB741266 KB631940 AEE63453.1 ENN71491.1 ERL87341.1 GEZM01038611 JAV81594.1 GDAI01002623 JAI14980.1 NEVH01000250 PNF43920.1 GFDF01006585 JAV07499.1 JXJN01000330 NEVH01005291 PNF38824.1 CCAG010018974 EZ423126 ADD19402.1 GDHF01010830 JAI41484.1 GAKP01001268 JAC57684.1 UFQS01000606 UFQT01000606 SSX05297.1 SSX25658.1 PYGN01000360 PSN47700.1 GBXI01010073 JAD04219.1 PYGN01000059 PSN55805.1 JRES01000644 KNC29663.1 KA644489 AFP59118.1 GECZ01015171 JAS54598.1 GAMC01003722 JAC02834.1 ADTU01024724 CH902619 EDV37534.1 CM000158 EDW91001.1 CH954179 EDV56053.1 CH480816 EDW47835.1 CM000362 CM002911 EDX07063.1 KMY93736.1 AJ000672 U91994 AE013599 AY095058 CH916367 EDW02076.1 CH940648 EDW62070.1 CH933808 EDW09511.1 CH479210 EDW32855.1 CM000071 EAL26461.1 KRT03116.1 CH963846 EDW72314.1 OUUW01000001 SPP74658.1 GAPW01001821 JAC11777.1 CVRI01000006 CRK88163.1 GANO01002165 JAB57706.1 CH477537 EAT39358.1 DS231829 EDS30323.1 GFDL01013926 JAV21119.1 AXCN02000699 AXCM01002571 AAAB01008964 EAA12847.3 APCN01002493 ATLV01023800 KE525346 KFB49604.1 GAHY01000478 JAA77032.1 GGFJ01006139 MBW55280.1 ADMH02000571 ETN65840.1 GGFK01010611 MBW43932.1 GEDC01031265 GEDC01026626 GEDC01011344 GEDC01006557 GEDC01003187 JAS06033.1 JAS10672.1 JAS25954.1 JAS30741.1 JAS34111.1 GGMR01015264 MBY27883.1 GFXV01001066 MBW12871.1 GGMS01002329 MBY71532.1 KZ288215 PBC32698.1 ABLF02024017 GEDC01024994 JAS12304.1 JF508489 AEB41038.1 DS235051 EEB10999.1 GDHC01012202 JAQ06427.1 GBRD01015862 JAG49964.1 GBHO01002788 JAG40816.1 GL762419 EFZ21587.1 QOIP01000007 RLU21064.1 KQ977171 KYN05137.1 KQ981241 KYN44308.1 GDIQ01002142 JAN92595.1 GBYB01003361 JAG73128.1 KQ979602 KYN20528.1
JAA92471.1 KQ460949 KPJ10457.1 KQ459185 KPJ03216.1 JF505286 AED87507.1 JTDY01000575 KOB76611.1 ODYU01000021 SOQ34082.1 FJ550361 NWSH01000825 ACR07790.1 PCG73997.1 KQ971351 EFA06522.1 APGK01055449 BT128496 KB741266 KB631940 AEE63453.1 ENN71491.1 ERL87341.1 GEZM01038611 JAV81594.1 GDAI01002623 JAI14980.1 NEVH01000250 PNF43920.1 GFDF01006585 JAV07499.1 JXJN01000330 NEVH01005291 PNF38824.1 CCAG010018974 EZ423126 ADD19402.1 GDHF01010830 JAI41484.1 GAKP01001268 JAC57684.1 UFQS01000606 UFQT01000606 SSX05297.1 SSX25658.1 PYGN01000360 PSN47700.1 GBXI01010073 JAD04219.1 PYGN01000059 PSN55805.1 JRES01000644 KNC29663.1 KA644489 AFP59118.1 GECZ01015171 JAS54598.1 GAMC01003722 JAC02834.1 ADTU01024724 CH902619 EDV37534.1 CM000158 EDW91001.1 CH954179 EDV56053.1 CH480816 EDW47835.1 CM000362 CM002911 EDX07063.1 KMY93736.1 AJ000672 U91994 AE013599 AY095058 CH916367 EDW02076.1 CH940648 EDW62070.1 CH933808 EDW09511.1 CH479210 EDW32855.1 CM000071 EAL26461.1 KRT03116.1 CH963846 EDW72314.1 OUUW01000001 SPP74658.1 GAPW01001821 JAC11777.1 CVRI01000006 CRK88163.1 GANO01002165 JAB57706.1 CH477537 EAT39358.1 DS231829 EDS30323.1 GFDL01013926 JAV21119.1 AXCN02000699 AXCM01002571 AAAB01008964 EAA12847.3 APCN01002493 ATLV01023800 KE525346 KFB49604.1 GAHY01000478 JAA77032.1 GGFJ01006139 MBW55280.1 ADMH02000571 ETN65840.1 GGFK01010611 MBW43932.1 GEDC01031265 GEDC01026626 GEDC01011344 GEDC01006557 GEDC01003187 JAS06033.1 JAS10672.1 JAS25954.1 JAS30741.1 JAS34111.1 GGMR01015264 MBY27883.1 GFXV01001066 MBW12871.1 GGMS01002329 MBY71532.1 KZ288215 PBC32698.1 ABLF02024017 GEDC01024994 JAS12304.1 JF508489 AEB41038.1 DS235051 EEB10999.1 GDHC01012202 JAQ06427.1 GBRD01015862 JAG49964.1 GBHO01002788 JAG40816.1 GL762419 EFZ21587.1 QOIP01000007 RLU21064.1 KQ977171 KYN05137.1 KQ981241 KYN44308.1 GDIQ01002142 JAN92595.1 GBYB01003361 JAG73128.1 KQ979602 KYN20528.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000037510
UP000218220
+ More
UP000007266 UP000192223 UP000019118 UP000030742 UP000235965 UP000092443 UP000092460 UP000091820 UP000078200 UP000092445 UP000092444 UP000245037 UP000095300 UP000037069 UP000095301 UP000005205 UP000007801 UP000192221 UP000002282 UP000008711 UP000001292 UP000000304 UP000000803 UP000001070 UP000008792 UP000009192 UP000008744 UP000001819 UP000007798 UP000079169 UP000268350 UP000183832 UP000008820 UP000002320 UP000075880 UP000075900 UP000075886 UP000075920 UP000075883 UP000076408 UP000075884 UP000075901 UP000007062 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000075840 UP000075885 UP000030765 UP000069272 UP000000673 UP000242457 UP000007819 UP000009046 UP000279307 UP000078542 UP000078541 UP000078492
UP000007266 UP000192223 UP000019118 UP000030742 UP000235965 UP000092443 UP000092460 UP000091820 UP000078200 UP000092445 UP000092444 UP000245037 UP000095300 UP000037069 UP000095301 UP000005205 UP000007801 UP000192221 UP000002282 UP000008711 UP000001292 UP000000304 UP000000803 UP000001070 UP000008792 UP000009192 UP000008744 UP000001819 UP000007798 UP000079169 UP000268350 UP000183832 UP000008820 UP000002320 UP000075880 UP000075900 UP000075886 UP000075920 UP000075883 UP000076408 UP000075884 UP000075901 UP000007062 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000075840 UP000075885 UP000030765 UP000069272 UP000000673 UP000242457 UP000007819 UP000009046 UP000279307 UP000078542 UP000078541 UP000078492
Interpro
IPR004536
SPS/SelD
+ More
IPR016188 PurM-like_N
IPR036676 PurM-like_C_sf
IPR036921 PurM-like_N_sf
IPR023061 SelD_I
IPR010918 PurM-like_C_dom
IPR036193 ADK_active_lid_dom_sf
IPR006259 Adenyl_kin_sub
IPR028586 AK3/Ak4_mitochondrial
IPR007862 Adenylate_kinase_lid-dom
IPR000850 Adenylat/UMP-CMP_kin
IPR027417 P-loop_NTPase
IPR033690 Adenylat_kinase_CS
IPR016188 PurM-like_N
IPR036676 PurM-like_C_sf
IPR036921 PurM-like_N_sf
IPR023061 SelD_I
IPR010918 PurM-like_C_dom
IPR036193 ADK_active_lid_dom_sf
IPR006259 Adenyl_kin_sub
IPR028586 AK3/Ak4_mitochondrial
IPR007862 Adenylate_kinase_lid-dom
IPR000850 Adenylat/UMP-CMP_kin
IPR027417 P-loop_NTPase
IPR033690 Adenylat_kinase_CS
Gene 3D
ProteinModelPortal
H9J385
Q3HR35
A0A212FNP3
S4PIS4
A0A194QYB3
A0A194QID8
+ More
F5CBM5 A0A0L7LN24 A0A2H1V151 C4NFK8 D6WQY1 A0A1W4XPA8 J3JZ15 A0A1Y1M7B0 A0A0K8TM22 A0A2J7RSV7 A0A1L8DM15 A0A1A9XPQ8 A0A1B0AM41 A0A1A9WIA8 A0A1A9V7P8 A0A2J7RDB1 A0A1A9ZPC6 D3TNX4 A0A0K8VRH3 A0A034WST6 A0A336KLL5 A0A2P8YTY2 A0A1I8PVW5 A0A0A1WYS7 A0A2P8ZH28 A0A0L0CB70 T1P7N8 A0A1B6FWM1 W8C155 A0A158NS94 B3MDY9 A0A1W4WBB6 B4P736 B3NR64 B4HRA6 B4QFF3 O18373 B4J758 B4LMH5 B4KSK6 B4H519 Q28X55 B4MJM5 A0A3Q0JDN0 A0A3B0JUA7 A0A023EQC1 A0A1J1HJS2 A0A1S3DEJ9 U5EUU9 Q16XK1 B0W2W4 A0A1Q3F0R0 A0A182IXD8 A0A182RJB0 A0A182QEQ1 A0A182W7I0 A0A182M2G5 A0A182XZU5 A0A182NTJ7 A0A182T8H0 Q7Q3N9 A0A1S4GWU2 A0A182USB2 A0A182JWN4 A0A182TUC4 A0A182WT47 A0A182L1Z5 A0A182HV42 A0A182PD02 A0A084WHB0 A0A182F301 R4FNR1 A0A2M4BQD9 W5JSF0 A0A2M4AT88 A0A1B6DYK4 A0A2S2PEK6 A0A2H8TFN3 A0A2S2Q2W3 A0A2A3ELU8 J9K9H8 A0A1B6CFT7 F5CBP8 E0VCB3 A0A146LHN0 A0A0K8S9W8 A0A0A9ZA82 E9ICW7 A0A3L8DKU4 A0A195CX57 A0A195FV21 A0A0P6JZ99 A0A0C9R999 A0A151J815
F5CBM5 A0A0L7LN24 A0A2H1V151 C4NFK8 D6WQY1 A0A1W4XPA8 J3JZ15 A0A1Y1M7B0 A0A0K8TM22 A0A2J7RSV7 A0A1L8DM15 A0A1A9XPQ8 A0A1B0AM41 A0A1A9WIA8 A0A1A9V7P8 A0A2J7RDB1 A0A1A9ZPC6 D3TNX4 A0A0K8VRH3 A0A034WST6 A0A336KLL5 A0A2P8YTY2 A0A1I8PVW5 A0A0A1WYS7 A0A2P8ZH28 A0A0L0CB70 T1P7N8 A0A1B6FWM1 W8C155 A0A158NS94 B3MDY9 A0A1W4WBB6 B4P736 B3NR64 B4HRA6 B4QFF3 O18373 B4J758 B4LMH5 B4KSK6 B4H519 Q28X55 B4MJM5 A0A3Q0JDN0 A0A3B0JUA7 A0A023EQC1 A0A1J1HJS2 A0A1S3DEJ9 U5EUU9 Q16XK1 B0W2W4 A0A1Q3F0R0 A0A182IXD8 A0A182RJB0 A0A182QEQ1 A0A182W7I0 A0A182M2G5 A0A182XZU5 A0A182NTJ7 A0A182T8H0 Q7Q3N9 A0A1S4GWU2 A0A182USB2 A0A182JWN4 A0A182TUC4 A0A182WT47 A0A182L1Z5 A0A182HV42 A0A182PD02 A0A084WHB0 A0A182F301 R4FNR1 A0A2M4BQD9 W5JSF0 A0A2M4AT88 A0A1B6DYK4 A0A2S2PEK6 A0A2H8TFN3 A0A2S2Q2W3 A0A2A3ELU8 J9K9H8 A0A1B6CFT7 F5CBP8 E0VCB3 A0A146LHN0 A0A0K8S9W8 A0A0A9ZA82 E9ICW7 A0A3L8DKU4 A0A195CX57 A0A195FV21 A0A0P6JZ99 A0A0C9R999 A0A151J815
PDB
3FD6
E-value=2.10176e-168,
Score=1520
Ontologies
KEGG
PATHWAY
GO
GO:0016260
GO:0005524
GO:0004756
GO:0005737
GO:0070329
GO:0001887
GO:0007005
GO:0006541
GO:0008284
GO:0007444
GO:0008283
GO:2000378
GO:0007049
GO:0016301
GO:0046033
GO:0005525
GO:0006172
GO:0046039
GO:0004017
GO:0046899
GO:0046041
GO:0005759
GO:0006355
GO:0008270
GO:0004828
GO:0006434
GO:0043564
GO:0006418
GO:0003824
PANTHER
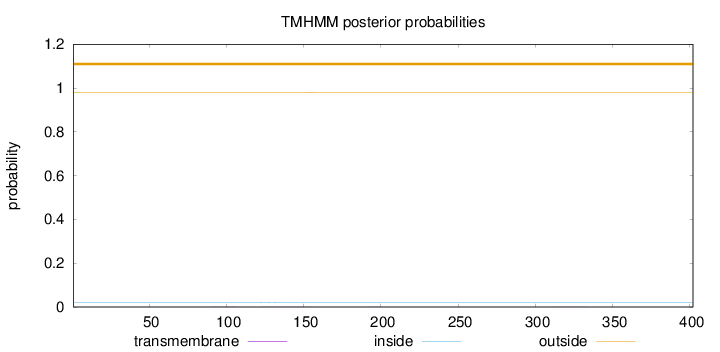
Topology
Subcellular location
Mitochondrion matrix
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0417100000000001
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.02170
outside
1 - 402
Population Genetic Test Statistics
Pi
213.679633
Theta
208.345833
Tajima's D
0.115702
CLR
0.225304
CSRT
0.402129893505325
Interpretation
Uncertain
