Pre Gene Modal
BGIBMGA004138
Annotation
PREDICTED:_activating_signal_cointegrator_1_[Papilio_machaon]
Full name
Molybdenum cofactor sulfurase
Alternative Name
Molybdenum cofactor sulfurtransferase
Protein maroon-like
Protein maroon-like
Location in the cell
Nuclear Reliability : 4.463
Sequence
CDS
ATGGAGCAATGGATAAGGGAAAATCTATCTACAATTTTGGATTTTGAAGTGCCTGACGATTTAATAAAGTATGTTTCGTCAATAGATAATGAAGTAGATTTAACTGAATACATGAAAACTTTGATTGACTTCAATAATTCTGAACATAAAAACTTTTTCTCCGAATTCATAAGGCTGAAGTTTCCTATTAAAGGCGGGACATCTAAACAGCAAAAAAAGAAAATATCAAAAGGCAAGGTTCAACAAGAGGTGATCGTCAAGGAAACAGTGCCTACACAAAGCGAGCCTGAAAGTAAAACTAAGAAGAAAACAAAATATGTCAATCTATACTCACAAGAAGGGAAGAATGCCCAAGTTGTGATGCTCAAAGGAAGACACCGCTGTGAATGTCAAGCTTCAAAACATGAGCTGATCAACAACTGCCTACAGTGCGGACGAGTGGTCTGCAGGCAGGAAGGCTCGGGCCCCTGTCTGTTTTGTGGCAGCTTGGTTTGCACTCCAGAAGAACAAAGGGAGTTGAATGCCAAGACCAAAGCTAGTGCTAAATTGATGGAGTCGCTTATGGAGAAGAGTCGACCCAAGGGCTGGGAGGACGCTCTGGTGCATAGAAATAGACTTTTGGAATATGATAGGACAAGTGAGCGTCGCACCCGCGTCACTGACGACGACAGTGACTACTTCAACGCGAACAGCGTGTGGCTCAGCTCAGAGGAGCGCGCCAAACTCGACAAGTACCAGCGGTCGCTGCACGAGAAGAAGCACGCGTCCAAACTGAACAAGAAGATGACCTTCGATTTCGCCGGGCGGCAGATATTGGAAGACAAAACCATAGACCACGATGTGGACGAGGACAAGATCAGAGACATAACCGCGGGAGCACGGGCCGCCGGCGGGGCGCTGCAGACGCACGCGGCCGTCGCCGGGCCAACGGACCGGGACGTGGCGCCCGGGGTCAACGCACCACTCTTACTGTTCGATCCGTCAGTGGAAAATGAGGATTATGCGACTGTTGTTAGAGCCAACGGGTTCTCCCAGATGTCTCGTATACAAGACGCCGAGTTGCAGGAGATGAGCGACACCGGCAGGTGCCTCTCCATGCACCAGCCCTGGGCCTCGTTGCTCGTTGAAGGCATCAAACTCCACGAAGGTCGTACATGGTACACCAGCCACCGAGGGAGGCTGTGGGTCGCCGCCACAGTGAAGCCCCCAGACCAGGACGTCATCAAGGCGCTGGAGAACCAGTACAGGGTCCTGTATCCAGAAAAGCAATTGCAATTTCCCTCGTTCTACCCGACTGGATGTCTGCTGGGCTGCGTCAACGTCGACGACTGCCTGCCCCAGGAGGAGTATCGGAAGACGTACCCGGACGGTGAGAGCGACAGTCCGTACGTGTTTATTTGCTCTAATCCTGTCAGCCTGAGGCTTCGGTTTCCGATCAAAGGACAACATAAAATATATCAACTAGAAAAAACAATTCATCAAGCCGCAATGAAATGTATTCAAAAAATGTCCAAAATACAAGCCAACGCAGCTTAA
Protein
MEQWIRENLSTILDFEVPDDLIKYVSSIDNEVDLTEYMKTLIDFNNSEHKNFFSEFIRLKFPIKGGTSKQQKKKISKGKVQQEVIVKETVPTQSEPESKTKKKTKYVNLYSQEGKNAQVVMLKGRHRCECQASKHELINNCLQCGRVVCRQEGSGPCLFCGSLVCTPEEQRELNAKTKASAKLMESLMEKSRPKGWEDALVHRNRLLEYDRTSERRTRVTDDDSDYFNANSVWLSSEERAKLDKYQRSLHEKKHASKLNKKMTFDFAGRQILEDKTIDHDVDEDKIRDITAGARAAGGALQTHAAVAGPTDRDVAPGVNAPLLLFDPSVENEDYATVVRANGFSQMSRIQDAELQEMSDTGRCLSMHQPWASLLVEGIKLHEGRTWYTSHRGRLWVAATVKPPDQDVIKALENQYRVLYPEKQLQFPSFYPTGCLLGCVNVDDCLPQEEYRKTYPDGESDSPYVFICSNPVSLRLRFPIKGQHKIYQLEKTIHQAAMKCIQKMSKIQANAA
Summary
Description
Sulfurates the molybdenum cofactor. Sulfation of molybdenum is essential for xanthine dehydrogenase (XDH) and aldehyde oxidase (ADO) enzymes in which molybdenum cofactor is liganded by 1 oxygen and 1 sulfur atom in active form.
Catalytic Activity
AH2 + L-cysteine + Mo-molybdopterin = A + H2O + L-alanine + thio-Mo-molybdopterin
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the class-V pyridoxal-phosphate-dependent aminotransferase family. MOCOS subfamily.
Uniprot
H9J3Q1
A0A2H1VUL3
A0A2A4JRA8
A0A194QCS3
A0A1E1WTC4
A0A212FNJ3
+ More
U5EU27 A0A182HFE2 A0A0K8TM87 Q16GG8 Q16P92 A0A1S4G1T0 A0A182QNA8 B0WQK0 A0A182Y7J0 A0A1Q3F612 A0A1Q3F4N2 A0A1Q3F3Z3 A0A084WQV2 A0A1Q3F418 A0A182LZA3 A0A182W4P4 A0A182JMU5 A0A182P7X1 A0A182N1H8 A0A182RBH3 A0A182K4H8 A0A2M4BJ98 A0A2M4BK05 A0A2M4BJ76 Q7PS17 W5JSK4 A0A182FEA0 A0A067QWU2 T1P8K6 A0A1I8MMJ2 A0A1I8QCR3 E0VGW2 A0A0K8U366 A0A034WHN0 A0A2J7R6K1 A0A0A1WJF3 A0A1J1HIQ0 A0A0L0C215 A0A1B6EYI7 K1R5U7 A0A0L7LPK0 A0A1A9VZM3 A0A1S3HGA7 E2BRE9 A0A1B6C1E5 A0A1B0FR62 A0A0L7RC76 A0A1W4VG59 R7T3K6 B3MW68 A0A2C9JJT8 A0A2C9JJT4 A0A0N0U590 B4PYH4 A0A2S2QIQ9 B3NY18 A0A2R2MMT0 A0A1B0C9F2 A0A1A9UEK6 A0A3S1B0D9 B4I684 Q9VRA1 A0A0B6ZWX8 A0A0C9R7G9 A0A182IA81 F7FX60 Q29GM2 A0A0M4ESR5 A0A3B0KKK2 A0A3M6TYR9 A0A2H8TGN7 A0A0B6ZW79 A0A182V472 G3VA38 J9JJC1 A0A0P5V261 A0A0N8D2Q7 A0A0N8AZN8 A0A0P4WD33 A0A336K089 A0A0P5WCT6 A0A210R4I4 A0A0P5XES7 A0A0A9XP76 A0A146LK62 A0A3Q3S910 A0A1B0ADQ0 B4NUT6 B4L337 A0A1B6C8K3 V4ABX4
U5EU27 A0A182HFE2 A0A0K8TM87 Q16GG8 Q16P92 A0A1S4G1T0 A0A182QNA8 B0WQK0 A0A182Y7J0 A0A1Q3F612 A0A1Q3F4N2 A0A1Q3F3Z3 A0A084WQV2 A0A1Q3F418 A0A182LZA3 A0A182W4P4 A0A182JMU5 A0A182P7X1 A0A182N1H8 A0A182RBH3 A0A182K4H8 A0A2M4BJ98 A0A2M4BK05 A0A2M4BJ76 Q7PS17 W5JSK4 A0A182FEA0 A0A067QWU2 T1P8K6 A0A1I8MMJ2 A0A1I8QCR3 E0VGW2 A0A0K8U366 A0A034WHN0 A0A2J7R6K1 A0A0A1WJF3 A0A1J1HIQ0 A0A0L0C215 A0A1B6EYI7 K1R5U7 A0A0L7LPK0 A0A1A9VZM3 A0A1S3HGA7 E2BRE9 A0A1B6C1E5 A0A1B0FR62 A0A0L7RC76 A0A1W4VG59 R7T3K6 B3MW68 A0A2C9JJT8 A0A2C9JJT4 A0A0N0U590 B4PYH4 A0A2S2QIQ9 B3NY18 A0A2R2MMT0 A0A1B0C9F2 A0A1A9UEK6 A0A3S1B0D9 B4I684 Q9VRA1 A0A0B6ZWX8 A0A0C9R7G9 A0A182IA81 F7FX60 Q29GM2 A0A0M4ESR5 A0A3B0KKK2 A0A3M6TYR9 A0A2H8TGN7 A0A0B6ZW79 A0A182V472 G3VA38 J9JJC1 A0A0P5V261 A0A0N8D2Q7 A0A0N8AZN8 A0A0P4WD33 A0A336K089 A0A0P5WCT6 A0A210R4I4 A0A0P5XES7 A0A0A9XP76 A0A146LK62 A0A3Q3S910 A0A1B0ADQ0 B4NUT6 B4L337 A0A1B6C8K3 V4ABX4
EC Number
2.8.1.9
Pubmed
19121390
26354079
22118469
26483478
26369729
17510324
+ More
25244985 24438588 12364791 14747013 17210077 20920257 23761445 24845553 25315136 20566863 25348373 25830018 26108605 22992520 26227816 20798317 23254933 17994087 15562597 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17495919 15632085 30382153 21709235 28812685 25401762 26823975
25244985 24438588 12364791 14747013 17210077 20920257 23761445 24845553 25315136 20566863 25348373 25830018 26108605 22992520 26227816 20798317 23254933 17994087 15562597 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17495919 15632085 30382153 21709235 28812685 25401762 26823975
EMBL
BABH01021739
ODYU01004536
SOQ44525.1
NWSH01000825
PCG73993.1
KQ459185
+ More
KPJ03219.1 GDQN01000867 JAT90187.1 AGBW02005277 OWR55314.1 GANO01001664 JAB58207.1 JXUM01134079 KQ568069 KXJ69154.1 GDAI01002134 JAI15469.1 CH478278 EAT33331.1 CH477791 EAT36176.1 AXCN02001394 DS232041 EDS32875.1 GFDL01012137 JAV22908.1 GFDL01012566 JAV22479.1 GFDL01012770 JAV22275.1 ATLV01025759 KE525399 KFB52596.1 GFDL01012762 JAV22283.1 AXCM01001655 GGFJ01003986 MBW53127.1 GGFJ01003987 MBW53128.1 GGFJ01003985 MBW53126.1 AAAB01008846 EAA06293.6 ADMH02000584 ETN65739.1 KK853153 KDR10551.1 KA644994 AFP59623.1 DS235152 EEB12618.1 GDHF01031326 JAI20988.1 GAKP01003861 JAC55091.1 NEVH01006756 PNF36465.1 GBXI01015108 JAC99183.1 CVRI01000004 CRK87434.1 JRES01000996 KNC26311.1 GECZ01026868 JAS42901.1 JH817887 EKC41083.1 JTDY01000400 KOB77357.1 GL449962 EFN81727.1 GEDC01029967 JAS07331.1 CCAG010018611 KQ414615 KOC68597.1 AMQN01003670 KB312450 ELT87196.1 CH902625 EDV35213.1 KQ435791 KOX74371.1 CM000162 EDX03015.1 GGMS01008432 MBY77635.1 CH954180 EDV47539.1 AJWK01002278 RQTK01001159 RUS71782.1 CH480823 EDW56290.1 AE014298 AAF50902.1 AHN59962.1 HACG01025987 CEK72852.1 GBYB01002711 JAG72478.1 APCN01001132 CH379064 EAL32087.1 CP012528 ALC49402.1 OUUW01000011 SPP86316.1 RCHS01002679 RMX46532.1 GFXV01001450 MBW13255.1 HACG01025988 CEK72853.1 AEFK01007746 AEFK01007747 AEFK01007748 ABLF02018136 GDIP01105937 JAL97777.1 GDIP01074591 LRGB01000115 JAM29124.1 KZS20794.1 GDIQ01227155 JAK24570.1 GDRN01061753 JAI65171.1 UFQS01000005 UFQT01000005 SSW96915.1 SSX17302.1 GDIP01088710 JAM15005.1 NEDP02000447 OWF55937.1 GDIP01073346 JAM30369.1 GBHO01024694 JAG18910.1 GDHC01010770 JAQ07859.1 CH984241 EDX16367.1 CH933810 EDW06965.1 GEDC01027461 JAS09837.1 KB202050 ESO92590.1
KPJ03219.1 GDQN01000867 JAT90187.1 AGBW02005277 OWR55314.1 GANO01001664 JAB58207.1 JXUM01134079 KQ568069 KXJ69154.1 GDAI01002134 JAI15469.1 CH478278 EAT33331.1 CH477791 EAT36176.1 AXCN02001394 DS232041 EDS32875.1 GFDL01012137 JAV22908.1 GFDL01012566 JAV22479.1 GFDL01012770 JAV22275.1 ATLV01025759 KE525399 KFB52596.1 GFDL01012762 JAV22283.1 AXCM01001655 GGFJ01003986 MBW53127.1 GGFJ01003987 MBW53128.1 GGFJ01003985 MBW53126.1 AAAB01008846 EAA06293.6 ADMH02000584 ETN65739.1 KK853153 KDR10551.1 KA644994 AFP59623.1 DS235152 EEB12618.1 GDHF01031326 JAI20988.1 GAKP01003861 JAC55091.1 NEVH01006756 PNF36465.1 GBXI01015108 JAC99183.1 CVRI01000004 CRK87434.1 JRES01000996 KNC26311.1 GECZ01026868 JAS42901.1 JH817887 EKC41083.1 JTDY01000400 KOB77357.1 GL449962 EFN81727.1 GEDC01029967 JAS07331.1 CCAG010018611 KQ414615 KOC68597.1 AMQN01003670 KB312450 ELT87196.1 CH902625 EDV35213.1 KQ435791 KOX74371.1 CM000162 EDX03015.1 GGMS01008432 MBY77635.1 CH954180 EDV47539.1 AJWK01002278 RQTK01001159 RUS71782.1 CH480823 EDW56290.1 AE014298 AAF50902.1 AHN59962.1 HACG01025987 CEK72852.1 GBYB01002711 JAG72478.1 APCN01001132 CH379064 EAL32087.1 CP012528 ALC49402.1 OUUW01000011 SPP86316.1 RCHS01002679 RMX46532.1 GFXV01001450 MBW13255.1 HACG01025988 CEK72853.1 AEFK01007746 AEFK01007747 AEFK01007748 ABLF02018136 GDIP01105937 JAL97777.1 GDIP01074591 LRGB01000115 JAM29124.1 KZS20794.1 GDIQ01227155 JAK24570.1 GDRN01061753 JAI65171.1 UFQS01000005 UFQT01000005 SSW96915.1 SSX17302.1 GDIP01088710 JAM15005.1 NEDP02000447 OWF55937.1 GDIP01073346 JAM30369.1 GBHO01024694 JAG18910.1 GDHC01010770 JAQ07859.1 CH984241 EDX16367.1 CH933810 EDW06965.1 GEDC01027461 JAS09837.1 KB202050 ESO92590.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000069940
UP000249989
+ More
UP000008820 UP000075886 UP000002320 UP000076408 UP000030765 UP000075883 UP000075920 UP000075880 UP000075885 UP000075884 UP000075900 UP000075881 UP000007062 UP000000673 UP000069272 UP000027135 UP000095301 UP000095300 UP000009046 UP000235965 UP000183832 UP000037069 UP000005408 UP000037510 UP000091820 UP000085678 UP000008237 UP000092444 UP000053825 UP000192221 UP000014760 UP000007801 UP000076420 UP000053105 UP000002282 UP000008711 UP000092461 UP000078200 UP000271974 UP000001292 UP000000803 UP000075840 UP000002280 UP000001819 UP000092553 UP000268350 UP000275408 UP000075903 UP000007648 UP000007819 UP000076858 UP000242188 UP000261640 UP000092445 UP000000304 UP000009192 UP000030746
UP000008820 UP000075886 UP000002320 UP000076408 UP000030765 UP000075883 UP000075920 UP000075880 UP000075885 UP000075884 UP000075900 UP000075881 UP000007062 UP000000673 UP000069272 UP000027135 UP000095301 UP000095300 UP000009046 UP000235965 UP000183832 UP000037069 UP000005408 UP000037510 UP000091820 UP000085678 UP000008237 UP000092444 UP000053825 UP000192221 UP000014760 UP000007801 UP000076420 UP000053105 UP000002282 UP000008711 UP000092461 UP000078200 UP000271974 UP000001292 UP000000803 UP000075840 UP000002280 UP000001819 UP000092553 UP000268350 UP000275408 UP000075903 UP000007648 UP000007819 UP000076858 UP000242188 UP000261640 UP000092445 UP000000304 UP000009192 UP000030746
Pfam
Interpro
IPR009349
Znf_C2HC5
+ More
IPR007374 ASCH_domain
IPR039128 TRIP4-like
IPR015947 PUA-like_sf
IPR015421 PyrdxlP-dep_Trfase_major
IPR000192 Aminotrans_V_dom
IPR005303 MOSC_N
IPR015424 PyrdxlP-dep_Trfase
IPR028886 MoCo_sulfurase
IPR005302 MoCF_Sase_C
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR011037 Pyrv_Knase-like_insert_dom_sf
IPR013134 Zn_hook_RAD50
IPR004584 Rad50_eukaryotes
IPR027417 P-loop_NTPase
IPR038729 Rad50/SbcC_AAA
IPR007374 ASCH_domain
IPR039128 TRIP4-like
IPR015947 PUA-like_sf
IPR015421 PyrdxlP-dep_Trfase_major
IPR000192 Aminotrans_V_dom
IPR005303 MOSC_N
IPR015424 PyrdxlP-dep_Trfase
IPR028886 MoCo_sulfurase
IPR005302 MoCF_Sase_C
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR011037 Pyrv_Knase-like_insert_dom_sf
IPR013134 Zn_hook_RAD50
IPR004584 Rad50_eukaryotes
IPR027417 P-loop_NTPase
IPR038729 Rad50/SbcC_AAA
Gene 3D
ProteinModelPortal
H9J3Q1
A0A2H1VUL3
A0A2A4JRA8
A0A194QCS3
A0A1E1WTC4
A0A212FNJ3
+ More
U5EU27 A0A182HFE2 A0A0K8TM87 Q16GG8 Q16P92 A0A1S4G1T0 A0A182QNA8 B0WQK0 A0A182Y7J0 A0A1Q3F612 A0A1Q3F4N2 A0A1Q3F3Z3 A0A084WQV2 A0A1Q3F418 A0A182LZA3 A0A182W4P4 A0A182JMU5 A0A182P7X1 A0A182N1H8 A0A182RBH3 A0A182K4H8 A0A2M4BJ98 A0A2M4BK05 A0A2M4BJ76 Q7PS17 W5JSK4 A0A182FEA0 A0A067QWU2 T1P8K6 A0A1I8MMJ2 A0A1I8QCR3 E0VGW2 A0A0K8U366 A0A034WHN0 A0A2J7R6K1 A0A0A1WJF3 A0A1J1HIQ0 A0A0L0C215 A0A1B6EYI7 K1R5U7 A0A0L7LPK0 A0A1A9VZM3 A0A1S3HGA7 E2BRE9 A0A1B6C1E5 A0A1B0FR62 A0A0L7RC76 A0A1W4VG59 R7T3K6 B3MW68 A0A2C9JJT8 A0A2C9JJT4 A0A0N0U590 B4PYH4 A0A2S2QIQ9 B3NY18 A0A2R2MMT0 A0A1B0C9F2 A0A1A9UEK6 A0A3S1B0D9 B4I684 Q9VRA1 A0A0B6ZWX8 A0A0C9R7G9 A0A182IA81 F7FX60 Q29GM2 A0A0M4ESR5 A0A3B0KKK2 A0A3M6TYR9 A0A2H8TGN7 A0A0B6ZW79 A0A182V472 G3VA38 J9JJC1 A0A0P5V261 A0A0N8D2Q7 A0A0N8AZN8 A0A0P4WD33 A0A336K089 A0A0P5WCT6 A0A210R4I4 A0A0P5XES7 A0A0A9XP76 A0A146LK62 A0A3Q3S910 A0A1B0ADQ0 B4NUT6 B4L337 A0A1B6C8K3 V4ABX4
U5EU27 A0A182HFE2 A0A0K8TM87 Q16GG8 Q16P92 A0A1S4G1T0 A0A182QNA8 B0WQK0 A0A182Y7J0 A0A1Q3F612 A0A1Q3F4N2 A0A1Q3F3Z3 A0A084WQV2 A0A1Q3F418 A0A182LZA3 A0A182W4P4 A0A182JMU5 A0A182P7X1 A0A182N1H8 A0A182RBH3 A0A182K4H8 A0A2M4BJ98 A0A2M4BK05 A0A2M4BJ76 Q7PS17 W5JSK4 A0A182FEA0 A0A067QWU2 T1P8K6 A0A1I8MMJ2 A0A1I8QCR3 E0VGW2 A0A0K8U366 A0A034WHN0 A0A2J7R6K1 A0A0A1WJF3 A0A1J1HIQ0 A0A0L0C215 A0A1B6EYI7 K1R5U7 A0A0L7LPK0 A0A1A9VZM3 A0A1S3HGA7 E2BRE9 A0A1B6C1E5 A0A1B0FR62 A0A0L7RC76 A0A1W4VG59 R7T3K6 B3MW68 A0A2C9JJT8 A0A2C9JJT4 A0A0N0U590 B4PYH4 A0A2S2QIQ9 B3NY18 A0A2R2MMT0 A0A1B0C9F2 A0A1A9UEK6 A0A3S1B0D9 B4I684 Q9VRA1 A0A0B6ZWX8 A0A0C9R7G9 A0A182IA81 F7FX60 Q29GM2 A0A0M4ESR5 A0A3B0KKK2 A0A3M6TYR9 A0A2H8TGN7 A0A0B6ZW79 A0A182V472 G3VA38 J9JJC1 A0A0P5V261 A0A0N8D2Q7 A0A0N8AZN8 A0A0P4WD33 A0A336K089 A0A0P5WCT6 A0A210R4I4 A0A0P5XES7 A0A0A9XP76 A0A146LK62 A0A3Q3S910 A0A1B0ADQ0 B4NUT6 B4L337 A0A1B6C8K3 V4ABX4
PDB
2E5O
E-value=5.44621e-44,
Score=448
Ontologies
GO
GO:0005634
GO:0006355
GO:0008270
GO:0003713
GO:0045893
GO:0016021
GO:0102867
GO:0030151
GO:0006777
GO:0030170
GO:0016829
GO:0008265
GO:0002020
GO:0035035
GO:0019901
GO:0005737
GO:0045661
GO:0044389
GO:0016604
GO:0030520
GO:0030331
GO:0005813
GO:0099053
GO:0031594
GO:1901998
GO:0016887
GO:0006281
GO:0030870
GO:0000723
GO:0005524
GO:0004828
GO:0006434
GO:0043564
GO:0006418
GO:0003824
PANTHER
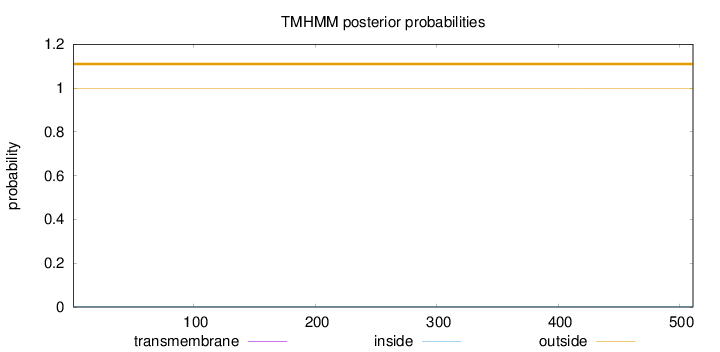
Topology
Length:
511
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00109
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00282
outside
1 - 511
Population Genetic Test Statistics
Pi
230.307196
Theta
183.84663
Tajima's D
1.168225
CLR
0.122825
CSRT
0.706114694265287
Interpretation
Uncertain
