Gene
KWMTBOMO11685 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003974
Annotation
NADH_dehydrogenase_[ubiquinone]_iron-sulfur_protein_5-like_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.291 Nuclear Reliability : 1.901
Sequence
CDS
ATGTCTTTGTCGCCGTTCTTTCGCTCACCGTTTACGGATGTTACTGGTGGCATGATTTCCCACCAACTTCTGGGTCGCTGCCAGAAAGAGGAAGCCAGATACATGGATTGTCTAGAAGCTTATGGTTTGGAACGAGGAAAGGTCAAGTGCGCTCATTTGTTCGGCGATTACCATGAATGCTCAACTCTTACCAAACAACTTAAGAGATTTTTGGCAATAAGACATGAGAGGCAGAGACAAATTTCTCAAGGAAAACTTACAGGCGACGAAAAGTATGTGAGCCCCCGTGTGGACAGTTACTAG
Protein
MSLSPFFRSPFTDVTGGMISHQLLGRCQKEEARYMDCLEAYGLERGKVKCAHLFGDYHECSTLTKQLKRFLAIRHERQRQISQGKLTGDEKYVSPRVDSY
Summary
Uniprot
H9J387
I4DJA5
A0A2A4JRE4
A0A1E1W6X9
A0A2H1VWD5
A0A194R3Q3
+ More
A0A212F879 S4PH09 H9J388 A0A0L7LIC6 A0A194QCT3 A0A3S2L5K1 A0A3S2NQX0 A0A2H1VUN4 A0A212F858 A0A0K8TPV2 B4LU70 B4MTU9 T1H6N3 A0A2P8YI02 A0A1I8PLW3 D6WSS5 A0A2J7R5F6 B3MUC9 A0A1W4UQT0 A0A182YDI1 A0A084WG33 A0A182RHA9 A0A182SBR2 Q29MZ9 B4G8F6 A0A2C9GR20 A0A182V0K2 A0A182K4A0 A0A182TFB2 A0A182KTU2 Q7Q1E9 B4JB75 A0A2M4A7T6 A0A2M4A7Y1 B3N783 W5J757 A0A2M4A884 A0A2M4A8C2 A0A1L8EEW0 A0A2M4CT70 B4ICS8 B4Q5N2 Q7K1C0 Q6XHP2 A0A182P312 A0A3B0K013 B4KHE9 A0A0L0C8G2 A0A2M3Z3V9 A0A2M4C4V4 A0A0A1WEN5 A0A182NC00 A0A182M7T3 A0A1A9WKV3 U5ER62 T1E3A6 A0A182R0K2 T1DFV9 B0W1B8 A0A1Q3FKM1 A0A182WGQ1 A0A182J1C8 A0A067RQI7 A0A023EER4 A0A2A4JPM8 A0A1Y1KBU5 A0A1W4XHU9 A0A182XGD5 W8BJJ8 Q1HRI5 A0A182F526 A0A1A9XFQ6 A0A1B0B7S7 A0A1A9UPZ1 A0A1I8N3D0 A0A0K8W277 A0A0C9RII6 A0A034VUV8 A0A1A9ZDF6 A0A1B0FDY5 A0A224XQN7 A0A336MC26 A0A1B0GKA9 A0A1L8E057 A0A1B6HT30 N6UCU1 U4UC75 A0A1Y1KT14 A0A0P4WPV6 A0A1B6FJI4 A0A224XQR1 A0A0A9XIK0 A0A1B6DI93 A0A2R7WN68
A0A212F879 S4PH09 H9J388 A0A0L7LIC6 A0A194QCT3 A0A3S2L5K1 A0A3S2NQX0 A0A2H1VUN4 A0A212F858 A0A0K8TPV2 B4LU70 B4MTU9 T1H6N3 A0A2P8YI02 A0A1I8PLW3 D6WSS5 A0A2J7R5F6 B3MUC9 A0A1W4UQT0 A0A182YDI1 A0A084WG33 A0A182RHA9 A0A182SBR2 Q29MZ9 B4G8F6 A0A2C9GR20 A0A182V0K2 A0A182K4A0 A0A182TFB2 A0A182KTU2 Q7Q1E9 B4JB75 A0A2M4A7T6 A0A2M4A7Y1 B3N783 W5J757 A0A2M4A884 A0A2M4A8C2 A0A1L8EEW0 A0A2M4CT70 B4ICS8 B4Q5N2 Q7K1C0 Q6XHP2 A0A182P312 A0A3B0K013 B4KHE9 A0A0L0C8G2 A0A2M3Z3V9 A0A2M4C4V4 A0A0A1WEN5 A0A182NC00 A0A182M7T3 A0A1A9WKV3 U5ER62 T1E3A6 A0A182R0K2 T1DFV9 B0W1B8 A0A1Q3FKM1 A0A182WGQ1 A0A182J1C8 A0A067RQI7 A0A023EER4 A0A2A4JPM8 A0A1Y1KBU5 A0A1W4XHU9 A0A182XGD5 W8BJJ8 Q1HRI5 A0A182F526 A0A1A9XFQ6 A0A1B0B7S7 A0A1A9UPZ1 A0A1I8N3D0 A0A0K8W277 A0A0C9RII6 A0A034VUV8 A0A1A9ZDF6 A0A1B0FDY5 A0A224XQN7 A0A336MC26 A0A1B0GKA9 A0A1L8E057 A0A1B6HT30 N6UCU1 U4UC75 A0A1Y1KT14 A0A0P4WPV6 A0A1B6FJI4 A0A224XQR1 A0A0A9XIK0 A0A1B6DI93 A0A2R7WN68
Pubmed
19121390
22651552
26354079
22118469
23622113
26227816
+ More
26369729 17994087 18057021 29403074 18362917 19820115 25244985 24438588 15632085 20966253 12364791 14747013 17210077 20920257 23761445 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14525923 17550304 26108605 25830018 24330624 24845553 24945155 26483478 28004739 24495485 17204158 17510324 25315136 25348373 23537049 25401762 26823975
26369729 17994087 18057021 29403074 18362917 19820115 25244985 24438588 15632085 20966253 12364791 14747013 17210077 20920257 23761445 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14525923 17550304 26108605 25830018 24330624 24845553 24945155 26483478 28004739 24495485 17204158 17510324 25315136 25348373 23537049 25401762 26823975
EMBL
BABH01021739
AK401373
KQ459185
BAM17995.1
KPJ03230.1
NWSH01000825
+ More
PCG73992.1 GDQN01008347 JAT82707.1 ODYU01004536 SOQ44524.1 KQ460949 KPJ10471.1 AGBW02009781 OWR49931.1 GAIX01005975 JAA86585.1 BABH01021738 JTDY01000952 KOB75308.1 KPJ03229.1 RSAL01000136 RVE46229.1 RVE46228.1 SOQ44523.1 OWR49932.1 GDAI01001445 JAI16158.1 CH940649 EDW64057.1 KRF81400.1 CH963852 EDW75538.1 PYGN01000580 PSN43870.1 KQ971352 EFA07503.1 NEVH01006994 PNF36066.1 CH902624 EDV33458.1 ATLV01023436 KE525343 KFB49177.1 CH379060 EAL33544.1 CH479180 EDW28636.1 APCN01001735 AAAB01008980 EAA14312.2 CH920880 CH916368 EDV90327.1 EDW03967.1 GGFK01003532 MBW36853.1 GGFK01003585 MBW36906.1 CH954177 EDV57190.1 KQS69902.1 ADMH02002127 ETN58619.1 GGFK01003692 MBW37013.1 GGFK01003661 MBW36982.1 GFDG01001583 JAV17216.1 GGFL01004193 MBW68371.1 CH480829 EDW45354.1 CM000361 CM002910 EDX03182.1 KMY87251.1 AE014134 AY069186 BT081978 AAF51538.1 AAL39331.1 AAN10510.1 ACO53093.1 ACZ94133.1 ACZ94134.1 AFH03492.1 AGB92329.1 AGB92332.1 AGB92333.1 AY232141 CM000157 AAR10164.1 EDW87098.1 OUUW01000004 SPP78956.1 CH933807 EDW11213.1 JRES01000753 KNC28728.1 GGFM01002453 MBW23204.1 GGFJ01011030 MBW60171.1 GBXI01016818 JAC97473.1 AXCM01003376 GANO01003911 JAB55960.1 GALA01000014 JAA94838.1 AXCN02000695 GAMD01003006 JAA98584.1 DS231821 EDS44988.1 GFDL01006918 JAV28127.1 KK852527 KDR22009.1 JXUM01042041 JXUM01074476 GAPW01006429 KQ562831 KQ561308 JAC07169.1 KXJ75051.1 KXJ79044.1 PCG74007.1 GEZM01087005 JAV58919.1 GAMC01007628 GAMC01007625 JAB98927.1 DQ440109 CH477825 ABF18142.1 EAT35870.1 EAT35871.1 JXJN01009707 GDHF01007344 JAI44970.1 GBYB01008060 JAG77827.1 GAKP01013631 GAKP01013629 GAKP01013627 GAKP01013624 JAC45323.1 CCAG010021136 GFTR01001609 JAW14817.1 UFQS01000888 UFQT01000888 SSX07505.1 SSX27845.1 AJWK01028622 GFDF01001944 JAV12140.1 GECU01029859 JAS77847.1 APGK01028665 KB740694 ENN79485.1 KB632194 ERL89898.1 GEZM01074769 JAV64474.1 GDRN01052275 JAI66318.1 GECZ01019424 JAS50345.1 GFTR01001581 JAW14845.1 GBHO01023785 GBRD01009436 GDHC01007457 JAG19819.1 JAG56388.1 JAQ11172.1 GEDC01024609 GEDC01011894 JAS12689.1 JAS25404.1 KK855038 PTY20440.1
PCG73992.1 GDQN01008347 JAT82707.1 ODYU01004536 SOQ44524.1 KQ460949 KPJ10471.1 AGBW02009781 OWR49931.1 GAIX01005975 JAA86585.1 BABH01021738 JTDY01000952 KOB75308.1 KPJ03229.1 RSAL01000136 RVE46229.1 RVE46228.1 SOQ44523.1 OWR49932.1 GDAI01001445 JAI16158.1 CH940649 EDW64057.1 KRF81400.1 CH963852 EDW75538.1 PYGN01000580 PSN43870.1 KQ971352 EFA07503.1 NEVH01006994 PNF36066.1 CH902624 EDV33458.1 ATLV01023436 KE525343 KFB49177.1 CH379060 EAL33544.1 CH479180 EDW28636.1 APCN01001735 AAAB01008980 EAA14312.2 CH920880 CH916368 EDV90327.1 EDW03967.1 GGFK01003532 MBW36853.1 GGFK01003585 MBW36906.1 CH954177 EDV57190.1 KQS69902.1 ADMH02002127 ETN58619.1 GGFK01003692 MBW37013.1 GGFK01003661 MBW36982.1 GFDG01001583 JAV17216.1 GGFL01004193 MBW68371.1 CH480829 EDW45354.1 CM000361 CM002910 EDX03182.1 KMY87251.1 AE014134 AY069186 BT081978 AAF51538.1 AAL39331.1 AAN10510.1 ACO53093.1 ACZ94133.1 ACZ94134.1 AFH03492.1 AGB92329.1 AGB92332.1 AGB92333.1 AY232141 CM000157 AAR10164.1 EDW87098.1 OUUW01000004 SPP78956.1 CH933807 EDW11213.1 JRES01000753 KNC28728.1 GGFM01002453 MBW23204.1 GGFJ01011030 MBW60171.1 GBXI01016818 JAC97473.1 AXCM01003376 GANO01003911 JAB55960.1 GALA01000014 JAA94838.1 AXCN02000695 GAMD01003006 JAA98584.1 DS231821 EDS44988.1 GFDL01006918 JAV28127.1 KK852527 KDR22009.1 JXUM01042041 JXUM01074476 GAPW01006429 KQ562831 KQ561308 JAC07169.1 KXJ75051.1 KXJ79044.1 PCG74007.1 GEZM01087005 JAV58919.1 GAMC01007628 GAMC01007625 JAB98927.1 DQ440109 CH477825 ABF18142.1 EAT35870.1 EAT35871.1 JXJN01009707 GDHF01007344 JAI44970.1 GBYB01008060 JAG77827.1 GAKP01013631 GAKP01013629 GAKP01013627 GAKP01013624 JAC45323.1 CCAG010021136 GFTR01001609 JAW14817.1 UFQS01000888 UFQT01000888 SSX07505.1 SSX27845.1 AJWK01028622 GFDF01001944 JAV12140.1 GECU01029859 JAS77847.1 APGK01028665 KB740694 ENN79485.1 KB632194 ERL89898.1 GEZM01074769 JAV64474.1 GDRN01052275 JAI66318.1 GECZ01019424 JAS50345.1 GFTR01001581 JAW14845.1 GBHO01023785 GBRD01009436 GDHC01007457 JAG19819.1 JAG56388.1 JAQ11172.1 GEDC01024609 GEDC01011894 JAS12689.1 JAS25404.1 KK855038 PTY20440.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000037510
+ More
UP000283053 UP000008792 UP000007798 UP000015102 UP000245037 UP000095300 UP000007266 UP000235965 UP000007801 UP000192221 UP000076408 UP000030765 UP000075900 UP000075901 UP000001819 UP000008744 UP000075840 UP000075903 UP000075881 UP000075902 UP000075882 UP000007062 UP000001070 UP000008711 UP000000673 UP000001292 UP000000304 UP000000803 UP000002282 UP000075885 UP000268350 UP000009192 UP000037069 UP000075884 UP000075883 UP000091820 UP000075886 UP000002320 UP000075920 UP000075880 UP000027135 UP000069940 UP000249989 UP000192223 UP000076407 UP000008820 UP000069272 UP000092443 UP000092460 UP000078200 UP000095301 UP000092445 UP000092444 UP000092461 UP000019118 UP000030742
UP000283053 UP000008792 UP000007798 UP000015102 UP000245037 UP000095300 UP000007266 UP000235965 UP000007801 UP000192221 UP000076408 UP000030765 UP000075900 UP000075901 UP000001819 UP000008744 UP000075840 UP000075903 UP000075881 UP000075902 UP000075882 UP000007062 UP000001070 UP000008711 UP000000673 UP000001292 UP000000304 UP000000803 UP000002282 UP000075885 UP000268350 UP000009192 UP000037069 UP000075884 UP000075883 UP000091820 UP000075886 UP000002320 UP000075920 UP000075880 UP000027135 UP000069940 UP000249989 UP000192223 UP000076407 UP000008820 UP000069272 UP000092443 UP000092460 UP000078200 UP000095301 UP000092445 UP000092444 UP000092461 UP000019118 UP000030742
Interpro
ProteinModelPortal
H9J387
I4DJA5
A0A2A4JRE4
A0A1E1W6X9
A0A2H1VWD5
A0A194R3Q3
+ More
A0A212F879 S4PH09 H9J388 A0A0L7LIC6 A0A194QCT3 A0A3S2L5K1 A0A3S2NQX0 A0A2H1VUN4 A0A212F858 A0A0K8TPV2 B4LU70 B4MTU9 T1H6N3 A0A2P8YI02 A0A1I8PLW3 D6WSS5 A0A2J7R5F6 B3MUC9 A0A1W4UQT0 A0A182YDI1 A0A084WG33 A0A182RHA9 A0A182SBR2 Q29MZ9 B4G8F6 A0A2C9GR20 A0A182V0K2 A0A182K4A0 A0A182TFB2 A0A182KTU2 Q7Q1E9 B4JB75 A0A2M4A7T6 A0A2M4A7Y1 B3N783 W5J757 A0A2M4A884 A0A2M4A8C2 A0A1L8EEW0 A0A2M4CT70 B4ICS8 B4Q5N2 Q7K1C0 Q6XHP2 A0A182P312 A0A3B0K013 B4KHE9 A0A0L0C8G2 A0A2M3Z3V9 A0A2M4C4V4 A0A0A1WEN5 A0A182NC00 A0A182M7T3 A0A1A9WKV3 U5ER62 T1E3A6 A0A182R0K2 T1DFV9 B0W1B8 A0A1Q3FKM1 A0A182WGQ1 A0A182J1C8 A0A067RQI7 A0A023EER4 A0A2A4JPM8 A0A1Y1KBU5 A0A1W4XHU9 A0A182XGD5 W8BJJ8 Q1HRI5 A0A182F526 A0A1A9XFQ6 A0A1B0B7S7 A0A1A9UPZ1 A0A1I8N3D0 A0A0K8W277 A0A0C9RII6 A0A034VUV8 A0A1A9ZDF6 A0A1B0FDY5 A0A224XQN7 A0A336MC26 A0A1B0GKA9 A0A1L8E057 A0A1B6HT30 N6UCU1 U4UC75 A0A1Y1KT14 A0A0P4WPV6 A0A1B6FJI4 A0A224XQR1 A0A0A9XIK0 A0A1B6DI93 A0A2R7WN68
A0A212F879 S4PH09 H9J388 A0A0L7LIC6 A0A194QCT3 A0A3S2L5K1 A0A3S2NQX0 A0A2H1VUN4 A0A212F858 A0A0K8TPV2 B4LU70 B4MTU9 T1H6N3 A0A2P8YI02 A0A1I8PLW3 D6WSS5 A0A2J7R5F6 B3MUC9 A0A1W4UQT0 A0A182YDI1 A0A084WG33 A0A182RHA9 A0A182SBR2 Q29MZ9 B4G8F6 A0A2C9GR20 A0A182V0K2 A0A182K4A0 A0A182TFB2 A0A182KTU2 Q7Q1E9 B4JB75 A0A2M4A7T6 A0A2M4A7Y1 B3N783 W5J757 A0A2M4A884 A0A2M4A8C2 A0A1L8EEW0 A0A2M4CT70 B4ICS8 B4Q5N2 Q7K1C0 Q6XHP2 A0A182P312 A0A3B0K013 B4KHE9 A0A0L0C8G2 A0A2M3Z3V9 A0A2M4C4V4 A0A0A1WEN5 A0A182NC00 A0A182M7T3 A0A1A9WKV3 U5ER62 T1E3A6 A0A182R0K2 T1DFV9 B0W1B8 A0A1Q3FKM1 A0A182WGQ1 A0A182J1C8 A0A067RQI7 A0A023EER4 A0A2A4JPM8 A0A1Y1KBU5 A0A1W4XHU9 A0A182XGD5 W8BJJ8 Q1HRI5 A0A182F526 A0A1A9XFQ6 A0A1B0B7S7 A0A1A9UPZ1 A0A1I8N3D0 A0A0K8W277 A0A0C9RII6 A0A034VUV8 A0A1A9ZDF6 A0A1B0FDY5 A0A224XQN7 A0A336MC26 A0A1B0GKA9 A0A1L8E057 A0A1B6HT30 N6UCU1 U4UC75 A0A1Y1KT14 A0A0P4WPV6 A0A1B6FJI4 A0A224XQR1 A0A0A9XIK0 A0A1B6DI93 A0A2R7WN68
PDB
5GUP
E-value=7.7682e-06,
Score=111
Ontologies
PATHWAY
PANTHER
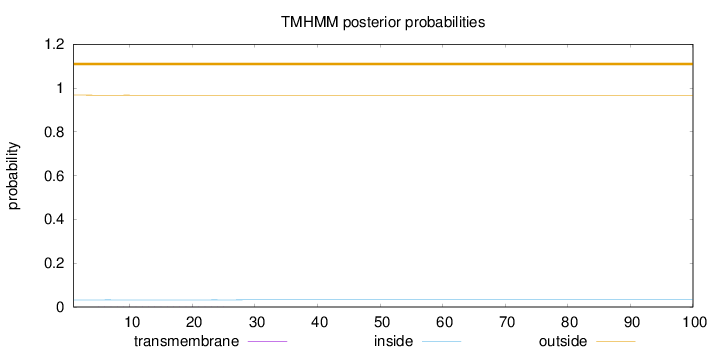
Topology
Length:
100
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00597
Exp number, first 60 AAs:
0.00597
Total prob of N-in:
0.03233
outside
1 - 100
Population Genetic Test Statistics
Pi
218.382669
Theta
187.701887
Tajima's D
0.514368
CLR
0.688645
CSRT
0.515674216289186
Interpretation
Uncertain
