Gene
KWMTBOMO11683
Pre Gene Modal
BGIBMGA003976
Annotation
tyrosine-protein_kinase_receptor_torso_[Bombyx_mori]
Full name
Tyrosine-protein kinase receptor torso
+ More
Fibroblast growth factor receptor
Fibroblast growth factor receptor 3
Fibroblast growth factor receptor
Fibroblast growth factor receptor 3
Alternative Name
PFR3
Location in the cell
Cytoplasmic Reliability : 2.067 Nuclear Reliability : 1.48
Sequence
CDS
ATGAAATTAAATAGCCGGTTCAATATTAAAAAACTCGATATATCCCCATTTTATATAATACAGGAAGACCCCGACGAGAAAACGCCCAAAGAAGACGATGTCGAAATCATCGGCATCGAATCGGGCAGCGCAGACGATCGCTGGGAGGTACGCTCGGACCGGGTGCTACTGCACGAAGTTATCGGCGAAGGAGCCTTCGGCGTCGTGCGGAGAGGCACTCTCGCGCCGGGAGGCAAGTCGGTCGCTGTTAAAATGTTAAAAGAATTTCCATCGCAGGAAGAAGTAAGGTCTTTTCGGTCTGAGATGGAGTTGATGAAGAGTGTAGGTGCTCATCCCCATGTTGTCAGTTTAGTTGGATGTTGTAGTGGACGAAAACCCCTCATTGTCGCTGAATATTGCAGTAGAGGAGATCTCCTCAGCTATTTGAGATGCTCTTGGGACATTATCGTGTCGAAACATACTGCAAAATACTACAATAATAATATGGACAGCATGGACACCTCCAAACTAAAAGTCCATAAAGAACACACAAAGCTTGTCGTTAACAAACTTTACGAGTTACAAGGTCCGTGTGAGACTGAACTAACGCCTTTAGACTTACTCTCTTTCTGTCGTCAAATCGCTATGGGCATGGAATTTTTAGCTGCAAACAGAATTGTACATCGGGACTTGGCTGCTAGAAATGTATTGGTAACTGAAGATAAGACTTTAAAGATAGCAGATTTTGGTCTATCTAGAGATATTTACGAAGAGAATCAGTATAAACAGAAAGGAAACGGTAAGATGCCTGTGAAATGGATGGCATTGGAGTCCTTGACTAGAAGGGTATACACGACGCAGAGTGATGTTTGGTCCTTCGGCGTGGTGATATGGGAGATAGTAACAGTGGGAGGCTCGCCATATCCAGAGGTGCCCGCAGCGCGCTTAGTGAGATCGTTAAGATCCGGTTACAGGATGCCAAAACCGGTCAACTGTAGTAAACCATTGTACGATATTATGAGGGCTTGTTGGAACGCGAGTCCTCGTGACAGGCCTACGTTTCCAGAGTTGCACCAGAAGCTCGACGACCTGCTGCATAGTGCGTGTGCAAACGAATATATAACTCTGGAAGTGGATGTTGACGAAGCACCTTCAACGCCGAAACCGCAAAGATACATCAAAATGTTGATCAGGGGCAAATTACCTTGGAGTCGTGAAAGTTACGAACGTCCTGTGAATCCAACAAGCAATCTGTACTCTTCGCCGCCGGTCATACAAACTAAAACCGCATGA
Protein
MKLNSRFNIKKLDISPFYIIQEDPDEKTPKEDDVEIIGIESGSADDRWEVRSDRVLLHEVIGEGAFGVVRRGTLAPGGKSVAVKMLKEFPSQEEVRSFRSEMELMKSVGAHPHVVSLVGCCSGRKPLIVAEYCSRGDLLSYLRCSWDIIVSKHTAKYYNNNMDSMDTSKLKVHKEHTKLVVNKLYELQGPCETELTPLDLLSFCRQIAMGMEFLAANRIVHRDLAARNVLVTEDKTLKIADFGLSRDIYEENQYKQKGNGKMPVKWMALESLTRRVYTTQSDVWSFGVVIWEIVTVGGSPYPEVPAARLVRSLRSGYRMPKPVNCSKPLYDIMRACWNASPRDRPTFPELHQKLDDLLHSACANEYITLEVDVDEAPSTPKPQRYIKMLIRGKLPWSRESYERPVNPTSNLYSSPPVIQTKTA
Summary
Description
Probable receptor tyrosine kinase. During postembryonic development, involved in the initiation of metamorphosis probably by inducing the production of ecdysone in response to prothoracicotropic hormone (PTTH) (By similarity). Binding to PTTH stimulates activation of canonical MAPK signaling leading to ERK phosphorylation (PubMed:19965758, PubMed:26928300).
Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation and apoptosis. Plays an essential role in the regulation of chondrocyte differentiation, proliferation and apoptosis, and is required for normal skeleton development. Regulates both osteogenesis and postnatal bone mineralization by osteoblasts. Promotes apoptosis in chondrocytes, but can also promote cancer cell proliferation. Phosphorylates PLCG1, CBL and FRS2. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway (By similarity).
Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation and apoptosis. Plays an essential role in the regulation of chondrocyte differentiation, proliferation and apoptosis, and is required for normal skeleton development. Regulates both osteogenesis and postnatal bone mineralization by osteoblasts. Promotes apoptosis in chondrocytes, but can also promote cancer cell proliferation. Phosphorylates PLCG1, CBL and FRS2. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway (By similarity).
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Cofactor
Mg(2+)
Subunit
Homodimer; disulfide-linked.
Monomer. Homodimer after ligand binding (By similarity).
Monomer. Homodimer after ligand binding (By similarity).
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family. Fibroblast growth factor receptor subfamily.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the protein kinase superfamily. Tyr protein kinase family. Fibroblast growth factor receptor subfamily.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Keywords
3D-structure
ATP-binding
Cell membrane
Complete proteome
Developmental protein
Disulfide bond
Glycoprotein
Kinase
Magnesium
Membrane
Metal-binding
Nucleotide-binding
Phosphoprotein
Reference proteome
Signal
Transferase
Transmembrane
Transmembrane helix
Tyrosine-protein kinase
Apoptosis
Immunoglobulin domain
Receptor
Repeat
Feature
chain Tyrosine-protein kinase receptor torso
Uniprot
D2IYS2
H9J389
A0A2H1VVK8
A0A0M3VGH2
A0A2A4JQG1
A0A3S2NFJ3
+ More
A0A212FKB6 A0A194QCN2 A0A194QZR5 A0A212F885 K7IR80 A0A151JAE2 E2C590 A0A158NVS0 A0A195FZ54 A0A151XEW5 A0A2P8ZBP9 A0A1Y1LTR3 E1ZXT5 F4X802 A0A151I268 A0A3L8DN84 A0A026WQS4 A0A067RFI1 A0A1B6FBG9 B0W484 D6WQC1 Q5DM52 A0A0U4HML1 Q16RK6 A0A1S4FS12 A7RHA0 A0A232EW20 A0A336MMX7 A0A182HF00 A0A147BNF1 A0A0V1F1G6 A0A0V1I7Q8 A0A0V1IAA9 A0A0V1N5B6 A0A146N2Q4 A0A0V1N629 A0A0V1N5K4 A0A0V1I8W2 A0A0V1JG26 A0A0V1N5R0 A0A0V1G383 A0A0V1JG69 A0A0V1K636 A0A0V0XXT6 A0A3M6TK71 A0A0V1K629 A0A2P2ICI7 A0A3B5L7D4 A3EXK4 A0A2G9S6I6 A0A0V1DEV4 A0A146N234 A0A0V1KQX4 A0A0V1DF45 A0A0V0VXZ8 A0A0V0VY18 A0A0V0SGB9 A0A0V1NNR9 A0A0V0TBE8 A0A0V1DF67 A0A2K6GP92 A0A0V1KPK4 A0A0V1NPE9 F6TEU5 A0A0V1AB90 A0A0V1C0M3 A0A0V1AB88 U3K5T0 A0A0V0SFD3 A0A0P7ULI3 A0A146N0E8 F1PK02 X1WJL7 A0A1Y3ED44 Q307W2 A0A3Q0GLC6 A0A3Q0GKJ3 A0A3L8SW15 K7FT64 Q91287 A0A2A4J3S4 Q5XUI1 V4AEI0 A0A0V0X9U6 A0A2A4J273 Q9U8W3 A0A2K6GPA8 A0A093PPX3 A0A0V0XAN3 I3JYB2 A0A093CYK4 A0A226EJW1 A0A0V0XA26 A0A340Y3U8
A0A212FKB6 A0A194QCN2 A0A194QZR5 A0A212F885 K7IR80 A0A151JAE2 E2C590 A0A158NVS0 A0A195FZ54 A0A151XEW5 A0A2P8ZBP9 A0A1Y1LTR3 E1ZXT5 F4X802 A0A151I268 A0A3L8DN84 A0A026WQS4 A0A067RFI1 A0A1B6FBG9 B0W484 D6WQC1 Q5DM52 A0A0U4HML1 Q16RK6 A0A1S4FS12 A7RHA0 A0A232EW20 A0A336MMX7 A0A182HF00 A0A147BNF1 A0A0V1F1G6 A0A0V1I7Q8 A0A0V1IAA9 A0A0V1N5B6 A0A146N2Q4 A0A0V1N629 A0A0V1N5K4 A0A0V1I8W2 A0A0V1JG26 A0A0V1N5R0 A0A0V1G383 A0A0V1JG69 A0A0V1K636 A0A0V0XXT6 A0A3M6TK71 A0A0V1K629 A0A2P2ICI7 A0A3B5L7D4 A3EXK4 A0A2G9S6I6 A0A0V1DEV4 A0A146N234 A0A0V1KQX4 A0A0V1DF45 A0A0V0VXZ8 A0A0V0VY18 A0A0V0SGB9 A0A0V1NNR9 A0A0V0TBE8 A0A0V1DF67 A0A2K6GP92 A0A0V1KPK4 A0A0V1NPE9 F6TEU5 A0A0V1AB90 A0A0V1C0M3 A0A0V1AB88 U3K5T0 A0A0V0SFD3 A0A0P7ULI3 A0A146N0E8 F1PK02 X1WJL7 A0A1Y3ED44 Q307W2 A0A3Q0GLC6 A0A3Q0GKJ3 A0A3L8SW15 K7FT64 Q91287 A0A2A4J3S4 Q5XUI1 V4AEI0 A0A0V0X9U6 A0A2A4J273 Q9U8W3 A0A2K6GPA8 A0A093PPX3 A0A0V0XAN3 I3JYB2 A0A093CYK4 A0A226EJW1 A0A0V0XA26 A0A340Y3U8
EC Number
2.7.10.1
Pubmed
19965758
26928300
26698662
19121390
26745499
22118469
+ More
26354079 20075255 20798317 21347285 29403074 28004739 21719571 30249741 24508170 24845553 18362917 19820115 16332539 17510324 17615350 28648823 26483478 29652888 30382153 17237944 17495919 16341006 30282656 17381049 8006062 16441300 23254933 10552041 25186727
26354079 20075255 20798317 21347285 29403074 28004739 21719571 30249741 24508170 24845553 18362917 19820115 16332539 17510324 17615350 28648823 26483478 29652888 30382153 17237944 17495919 16341006 30282656 17381049 8006062 16441300 23254933 10552041 25186727
EMBL
GQ477743
ACZ81657.1
BABH01021736
ODYU01004535
SOQ44522.1
LC076477
+ More
BAS29645.2 NWSH01000825 PCG74006.1 RSAL01000136 RVE46230.1 AGBW02008079 OWR54183.1 KQ459185 KPJ03227.1 KQ460949 KPJ10470.1 AGBW02009781 OWR49933.1 KQ979294 KYN22004.1 GL452767 EFN76911.1 ADTU01027399 ADTU01027400 ADTU01027401 KQ981193 KYN45094.1 KQ982249 KYQ58830.1 PYGN01000114 PSN53900.1 GEZM01050174 JAV75690.1 GL435089 EFN74006.1 GL888900 EGI57419.1 KQ976554 KYM80805.1 QOIP01000006 RLU21841.1 KK107128 EZA58397.1 KK852718 KDR17781.1 GECZ01022506 JAS47263.1 DS231835 EDS32928.1 KQ971354 EFA07525.1 AY618898 AAU87291.1 KR152228 ALX72397.1 CH477706 EAT37041.1 KU746914 DS469510 AOP31969.1 EDO49315.1 NNAY01001930 OXU22523.1 UFQT01001748 SSX31646.1 JXUM01133095 JXUM01133096 GEGO01003432 JAR91972.1 JYDR01000001 KRY79790.1 JYDP01000002 KRZ18899.1 KRZ18901.1 JYDO01000008 KRZ79179.1 GCES01160986 JAQ25336.1 KRZ79182.1 KRZ79183.1 KRZ18900.1 JYDS01000006 KRZ33929.1 KRZ79184.1 JYDT01000005 KRY92792.1 KRZ33930.1 JYDV01000013 KRZ42674.1 JYDU01000103 KRX92803.1 RCHS01003437 RMX41827.1 KRZ42675.1 IACF01006134 LAB71717.1 EF068147 ABN70838.1 KV926580 PIO35695.1 JYDI01000008 KRY60087.1 GCES01160985 JAQ25337.1 JYDW01000323 KRZ49262.1 KRY60090.1 JYDN01000002 KRX68375.1 KRX68376.1 JYDL01000012 KRX25317.1 JYDM01000136 KRZ85679.1 JYDJ01000370 KRX36351.1 KRY60085.1 KRZ49261.1 KRZ85680.1 JYDQ01000012 KRY22088.1 JYDH01000003 KRY42868.1 KRY22087.1 AGTO01010340 KRX25318.1 JARO02010852 KPP60354.1 GCES01160984 JAQ25338.1 AAEX03002461 ABLF02021638 ABLF02021640 ABLF02021643 ABLF02021646 ABLF02051162 LVZM01021957 OUC41048.1 DQ235692 ABB52005.1 QUSF01000005 RLW09234.1 AGCU01164154 AGCU01164155 AGCU01164156 AGCU01164157 AGCU01164158 AGCU01164159 AGCU01164160 AGCU01164161 AGCU01164162 X75603 NWSH01003526 PCG66164.1 AY737276 AAU89298.1 KB201891 ESO93545.1 JYDK01000005 KRX84680.1 PCG66165.1 AB025537 BAA84727.1 KL670101 KFW78496.1 KRX84675.1 AERX01013706 AERX01013707 KL468555 KFV17964.1 LNIX01000003 OXA57985.1 KRX84677.1
BAS29645.2 NWSH01000825 PCG74006.1 RSAL01000136 RVE46230.1 AGBW02008079 OWR54183.1 KQ459185 KPJ03227.1 KQ460949 KPJ10470.1 AGBW02009781 OWR49933.1 KQ979294 KYN22004.1 GL452767 EFN76911.1 ADTU01027399 ADTU01027400 ADTU01027401 KQ981193 KYN45094.1 KQ982249 KYQ58830.1 PYGN01000114 PSN53900.1 GEZM01050174 JAV75690.1 GL435089 EFN74006.1 GL888900 EGI57419.1 KQ976554 KYM80805.1 QOIP01000006 RLU21841.1 KK107128 EZA58397.1 KK852718 KDR17781.1 GECZ01022506 JAS47263.1 DS231835 EDS32928.1 KQ971354 EFA07525.1 AY618898 AAU87291.1 KR152228 ALX72397.1 CH477706 EAT37041.1 KU746914 DS469510 AOP31969.1 EDO49315.1 NNAY01001930 OXU22523.1 UFQT01001748 SSX31646.1 JXUM01133095 JXUM01133096 GEGO01003432 JAR91972.1 JYDR01000001 KRY79790.1 JYDP01000002 KRZ18899.1 KRZ18901.1 JYDO01000008 KRZ79179.1 GCES01160986 JAQ25336.1 KRZ79182.1 KRZ79183.1 KRZ18900.1 JYDS01000006 KRZ33929.1 KRZ79184.1 JYDT01000005 KRY92792.1 KRZ33930.1 JYDV01000013 KRZ42674.1 JYDU01000103 KRX92803.1 RCHS01003437 RMX41827.1 KRZ42675.1 IACF01006134 LAB71717.1 EF068147 ABN70838.1 KV926580 PIO35695.1 JYDI01000008 KRY60087.1 GCES01160985 JAQ25337.1 JYDW01000323 KRZ49262.1 KRY60090.1 JYDN01000002 KRX68375.1 KRX68376.1 JYDL01000012 KRX25317.1 JYDM01000136 KRZ85679.1 JYDJ01000370 KRX36351.1 KRY60085.1 KRZ49261.1 KRZ85680.1 JYDQ01000012 KRY22088.1 JYDH01000003 KRY42868.1 KRY22087.1 AGTO01010340 KRX25318.1 JARO02010852 KPP60354.1 GCES01160984 JAQ25338.1 AAEX03002461 ABLF02021638 ABLF02021640 ABLF02021643 ABLF02021646 ABLF02051162 LVZM01021957 OUC41048.1 DQ235692 ABB52005.1 QUSF01000005 RLW09234.1 AGCU01164154 AGCU01164155 AGCU01164156 AGCU01164157 AGCU01164158 AGCU01164159 AGCU01164160 AGCU01164161 AGCU01164162 X75603 NWSH01003526 PCG66164.1 AY737276 AAU89298.1 KB201891 ESO93545.1 JYDK01000005 KRX84680.1 PCG66165.1 AB025537 BAA84727.1 KL670101 KFW78496.1 KRX84675.1 AERX01013706 AERX01013707 KL468555 KFV17964.1 LNIX01000003 OXA57985.1 KRX84677.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000002358 UP000078492 UP000008237 UP000005205 UP000078541 UP000075809 UP000245037 UP000000311 UP000007755 UP000078540 UP000279307 UP000053097 UP000027135 UP000002320 UP000007266 UP000008820 UP000001593 UP000215335 UP000069940 UP000054632 UP000055024 UP000054843 UP000054805 UP000054995 UP000054826 UP000054815 UP000275408 UP000261380 UP000054653 UP000054721 UP000054681 UP000054630 UP000054924 UP000055048 UP000233160 UP000002280 UP000054783 UP000054776 UP000016665 UP000034805 UP000002254 UP000007819 UP000243006 UP000189705 UP000276834 UP000007267 UP000030746 UP000054673 UP000053258 UP000005207 UP000198287 UP000265300
UP000002358 UP000078492 UP000008237 UP000005205 UP000078541 UP000075809 UP000245037 UP000000311 UP000007755 UP000078540 UP000279307 UP000053097 UP000027135 UP000002320 UP000007266 UP000008820 UP000001593 UP000215335 UP000069940 UP000054632 UP000055024 UP000054843 UP000054805 UP000054995 UP000054826 UP000054815 UP000275408 UP000261380 UP000054653 UP000054721 UP000054681 UP000054630 UP000054924 UP000055048 UP000233160 UP000002280 UP000054783 UP000054776 UP000016665 UP000034805 UP000002254 UP000007819 UP000243006 UP000189705 UP000276834 UP000007267 UP000030746 UP000054673 UP000053258 UP000005207 UP000198287 UP000265300
Interpro
IPR008266
Tyr_kinase_AS
+ More
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR036116 FN3_sf
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR036179 Ig-like_dom_sf
IPR016248 FGF_rcpt_fam
IPR003598 Ig_sub2
IPR013106 Ig_V-set
IPR041159 FGFR_TM
IPR028174 FGF_rcpt_1
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR019821 Kinesin_motor_CS
IPR036961 Kinesin_motor_dom_sf
IPR013151 Immunoglobulin
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR036116 FN3_sf
IPR003961 FN3_dom
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR036179 Ig-like_dom_sf
IPR016248 FGF_rcpt_fam
IPR003598 Ig_sub2
IPR013106 Ig_V-set
IPR041159 FGFR_TM
IPR028174 FGF_rcpt_1
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR019821 Kinesin_motor_CS
IPR036961 Kinesin_motor_dom_sf
IPR013151 Immunoglobulin
Gene 3D
ProteinModelPortal
D2IYS2
H9J389
A0A2H1VVK8
A0A0M3VGH2
A0A2A4JQG1
A0A3S2NFJ3
+ More
A0A212FKB6 A0A194QCN2 A0A194QZR5 A0A212F885 K7IR80 A0A151JAE2 E2C590 A0A158NVS0 A0A195FZ54 A0A151XEW5 A0A2P8ZBP9 A0A1Y1LTR3 E1ZXT5 F4X802 A0A151I268 A0A3L8DN84 A0A026WQS4 A0A067RFI1 A0A1B6FBG9 B0W484 D6WQC1 Q5DM52 A0A0U4HML1 Q16RK6 A0A1S4FS12 A7RHA0 A0A232EW20 A0A336MMX7 A0A182HF00 A0A147BNF1 A0A0V1F1G6 A0A0V1I7Q8 A0A0V1IAA9 A0A0V1N5B6 A0A146N2Q4 A0A0V1N629 A0A0V1N5K4 A0A0V1I8W2 A0A0V1JG26 A0A0V1N5R0 A0A0V1G383 A0A0V1JG69 A0A0V1K636 A0A0V0XXT6 A0A3M6TK71 A0A0V1K629 A0A2P2ICI7 A0A3B5L7D4 A3EXK4 A0A2G9S6I6 A0A0V1DEV4 A0A146N234 A0A0V1KQX4 A0A0V1DF45 A0A0V0VXZ8 A0A0V0VY18 A0A0V0SGB9 A0A0V1NNR9 A0A0V0TBE8 A0A0V1DF67 A0A2K6GP92 A0A0V1KPK4 A0A0V1NPE9 F6TEU5 A0A0V1AB90 A0A0V1C0M3 A0A0V1AB88 U3K5T0 A0A0V0SFD3 A0A0P7ULI3 A0A146N0E8 F1PK02 X1WJL7 A0A1Y3ED44 Q307W2 A0A3Q0GLC6 A0A3Q0GKJ3 A0A3L8SW15 K7FT64 Q91287 A0A2A4J3S4 Q5XUI1 V4AEI0 A0A0V0X9U6 A0A2A4J273 Q9U8W3 A0A2K6GPA8 A0A093PPX3 A0A0V0XAN3 I3JYB2 A0A093CYK4 A0A226EJW1 A0A0V0XA26 A0A340Y3U8
A0A212FKB6 A0A194QCN2 A0A194QZR5 A0A212F885 K7IR80 A0A151JAE2 E2C590 A0A158NVS0 A0A195FZ54 A0A151XEW5 A0A2P8ZBP9 A0A1Y1LTR3 E1ZXT5 F4X802 A0A151I268 A0A3L8DN84 A0A026WQS4 A0A067RFI1 A0A1B6FBG9 B0W484 D6WQC1 Q5DM52 A0A0U4HML1 Q16RK6 A0A1S4FS12 A7RHA0 A0A232EW20 A0A336MMX7 A0A182HF00 A0A147BNF1 A0A0V1F1G6 A0A0V1I7Q8 A0A0V1IAA9 A0A0V1N5B6 A0A146N2Q4 A0A0V1N629 A0A0V1N5K4 A0A0V1I8W2 A0A0V1JG26 A0A0V1N5R0 A0A0V1G383 A0A0V1JG69 A0A0V1K636 A0A0V0XXT6 A0A3M6TK71 A0A0V1K629 A0A2P2ICI7 A0A3B5L7D4 A3EXK4 A0A2G9S6I6 A0A0V1DEV4 A0A146N234 A0A0V1KQX4 A0A0V1DF45 A0A0V0VXZ8 A0A0V0VY18 A0A0V0SGB9 A0A0V1NNR9 A0A0V0TBE8 A0A0V1DF67 A0A2K6GP92 A0A0V1KPK4 A0A0V1NPE9 F6TEU5 A0A0V1AB90 A0A0V1C0M3 A0A0V1AB88 U3K5T0 A0A0V0SFD3 A0A0P7ULI3 A0A146N0E8 F1PK02 X1WJL7 A0A1Y3ED44 Q307W2 A0A3Q0GLC6 A0A3Q0GKJ3 A0A3L8SW15 K7FT64 Q91287 A0A2A4J3S4 Q5XUI1 V4AEI0 A0A0V0X9U6 A0A2A4J273 Q9U8W3 A0A2K6GPA8 A0A093PPX3 A0A0V0XAN3 I3JYB2 A0A093CYK4 A0A226EJW1 A0A0V0XA26 A0A340Y3U8
PDB
4K33
E-value=6.20005e-73,
Score=697
Ontologies
GO
GO:0005524
GO:0005886
GO:0046872
GO:0004714
GO:0007275
GO:0016021
GO:0004713
GO:0022008
GO:0007399
GO:0043235
GO:0005887
GO:0030154
GO:0030424
GO:0043410
GO:0051897
GO:0007169
GO:0008284
GO:0005007
GO:0046777
GO:0005794
GO:0070374
GO:0010518
GO:0042531
GO:0017134
GO:1902178
GO:0042802
GO:0043552
GO:0005783
GO:0030133
GO:0006915
GO:0003777
GO:0007018
GO:0008017
GO:0006468
GO:0004672
GO:0004828
GO:0005737
GO:0006434
GO:0043564
GO:0006418
GO:0003824
Topology
Subcellular location
Cell membrane
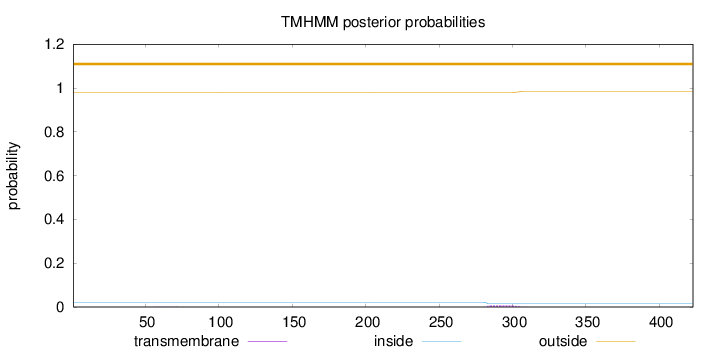
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15357
Exp number, first 60 AAs:
0.00077
Total prob of N-in:
0.02134
outside
1 - 423
Population Genetic Test Statistics
Pi
82.057074
Theta
58.902378
Tajima's D
0.887145
CLR
1.205889
CSRT
0.633918304084796
Interpretation
Uncertain
