Pre Gene Modal
BGIBMGA004137
Annotation
PREDICTED:_dnaJ_homolog_subfamily_C_member_13_isoform_X3_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.16 Nuclear Reliability : 1.317
Sequence
CDS
ATGATTCCACTAAAGGACAATCAGGATGTGGCATCGTTCCTTGTCACAAAACACTCTTGGAAGGGGAAGTACAAGAGAGTGTTTTCTATTGGAACTCATGGAATTACTACATACAATCCAGATCGCCTGGAAGTCACCAACAAATGGCTGTATGCTGATGTAGTTACCATTGCATCAGCAAAGCATTCAAACTCTACTGCCAACCATGACTTCACACTTGTCATGAAAAAAGAAAAGAAAGTGGACTCAATGCGATTTTCTTCAGAGCACAAGTGTTTGATACTCACTGAAGCTTTTAAATATAGGCATTTGTTTGCGGAGAAACCAAAGGATGTTTTTAGATACCAAGCATATAAACATCACTGGAGTGGAACAAGACTACCAATAGTACTTGAAGTTGGGCCTTGTGCTTTAGAACAGCTAGACCCATCGACACACACTCAGCTAGCCTTCTACCCATACTGTGATATACAGGGTATCCTTCCCGTACGAGATGTAGCTGGTGGTTTTATGCTTGCTGTTGGAGGTTATAGTCGCTTACATCTGTTTTCAAATGCAATGGATCACCAGATTATTATAAATAAGATGCTTGAATTAGCCAATTTGACATTGAGCATTAGCATCAAGGTTTTATCAGCTACAATTACATTGGATGATTATCATGATCAACGTTTTGGAAAATATAGTGGAGACCAACATCAAACATCGTTGAGCGAGTTCATCGTGCACAAAGTTAACCCAGCTCGTCATATGGAGCCCATGCGTCGTACACTCTGTCTTTCAGATACATGTATTCTAGAAAGAGATCCACAAACATACTCCGTTGTATGTTTGCGTCCTTTGAGCGACGTTTTTGCTTTGATAAGAGATCCTGAACAGCCTCAGAAATTCTCAATCGAATACATGAATGGACAGACGCGTACTTATTTAGCTGGCGAAAGGGACGCGTTATTGGCGTCGTTATTAGATGGTGTTCGTTCGGCGGGCCAGCGCGACGTGCACGTGCGTTCTACGTACACGCCGCGCGGTTACAGACTCGGTCCCTTACACCACAGCGTCGACGAGGAGACTGAATCTAATCATCTAAGATTGTTTCAAAATCCAGTTGGTATGACTCGCGCGGAAGTAATAGAACGTTTCAACGCCAACATCGGATACAGCGGTCTAATATACTCCGTTAGCCAAGACCGTTTGTTCGCGGAGAACAAAGAGAAGTTGATAACGGGCGCCCTGACCGCGCTGGTGAACAGTGTGAGCTCCAGCGCCACGCTGCCCCCGCGCGAGCTGGAGGCGCAGTTCCACGCGCTGCGGAGGCTCTGCGCCAGTAAAGTGGGCTTCGCTGCCTTCACAGCCTTGACCGGGTTCCGCGAGTGGGTGGGCGCGTCCGTGGTGGCGGCGCTGCGGCGCGGCAGTGCGGCGTGCTCGCACGCGGCGCTGGACGCGGTGTGCGCGCTCATGCAGCCCATGCACCACGAGCCCGACCTGCGCCAGGAGCAGCTCAACAAGGCCAGCATGCTCTCGTCCGCGCAGTTCCTCGACGGCCTGCTCGCCATGTGGGCGGAACACGTTAGTGCAGGCACCGGGGCTTTGGTAGTGGCAGCCATGTTGGATTTCTTAACGTTCGCCTTGTGCGTGCCGTATAGTGAAACCACGGATGGAAAACAGTTTGACCAGCTATTGGAAATGGTTTCTAATAAGGGACGCGTCATATTTAAATTGTTTGAACACCCGTCAATGGCAATAGTAAAAGGAGCAGGCCTCGTGATGAGAGCGTTGATCGAAGAGGGCGCCGGTGCCGTGGGCACTCGCATGCAGGCCCTCGCTCTGGCTGAGGCAGCCCTGCCGAGGCATCTCCTCACCGCGCTCTACGCTCCAGCGAGACTGCATCACAGGCATCTGGCGCGACATCTGGTGTCGCTGTGGTTGGTCGACCACGCACCGGCGCATAACCTCCTCAGCAGGATAATGCCGTCGGGGCTGCTCGGGTTCCTGGAGTCGGCGGAGAGTCCCCCGGCGGGCGCGCTGGAGGAGCAGCGGGACGAGGGGCGCGACAACCTGCAGCTGGCGCAGGCCGCGCATCGCCCGCGCGCCTCGCACTGGCAGGCGCTCGACCGACAGGTCAAGTACGTCGAGAAACATCTGGAGCACTACGCCAGCCTGGCGCTGCAGCACTGGGCGCAGCGCACGGGGCGCGCGGGGGCTGGGGCGGGGGGCGCGGCGCGGGAGGCGGTGCGGGAGCGGCCCGTGCTGTTGCGGCGGCGCCGGGAGCGGGTCAAGTCCGCCGCCAACTGGCCGCTGTTCTACTACCAGTTCCACAGGGACCACGCCCTGCCTAATCTCATTTGGAACCATACGACCCGGGAGGAGCTCCGTAACGCCCTGGAGAACGAGCTGCGCGCGTTCACTTCGGACCGCGAAGTGGCCGGCAACACTCTCACTTCGTGGAACCACGCCGAGCTGGAGCTGCAGTACCAGTGTCTCCAGAACGAAGTCAAGATCGGCGACTATTATCTAAGGATACTGCTGGAGCAGAAAGACAACGACGACAGCCCGATACGGAAGTCGTATGAGTTCTTTAACGACCTGTACCATCGGTTCCTGTCCACCCCCAAAGTGGAGATGAAGTGCATGTGCCTGCAGGCGATGAGCATCGTGTACGGGCGGTACTTCGAGGACATCGGACCTTTTGCCGACACCAGATACATCGTGCAGATGTTGGACAGGACCTGTGATAGAATGGAGAGAGATAGACTTGTTCAGTTTTTGTCGAAACTAATACTACACAAACGAAACGTTAGTGAGATATTAGAATGGAACGGAATTAGGATCTTAGTGGAGCTGATGACGTTGGCACACCTGCACACGTCGCGGGCGCTCATCCCGGCGCAGAGCAACGTCATCGAGGGCCCCGCCGAGCCCGGGGGCAACGACCGCGAGTGGTACTACAACCTCGAGCAGGGAGACAAGGTTCAGAGGAAGGGTCCCATCAGTCTTCAACAGCTAAAGGACCTGTACAAGTCTGGCGAGATCACGAACAAGACGAAATGTTGGGCCAACAGCATGGAAGGTTGGCGTGCAGTTGGCGGTATTCCCCAACTCAAGTGGACGCTGGTGGCGCGGGGAGCCCCAGTTTTCGATGAAAGCGCTCTGGCCGCTACGATTTTGGATCTATTGATTACTTGTTCACGATACTATCCGAGCAGAGACGAAGAAGACGCGGTAATAAGGCCGCTCCCAAGAATAAAGCGGCTGCTCTCGGAGCCGGCCTGTCTGGCGCACGTCGTCCAGTTGCTGCTGACCTTCGATCCCATACTCGTCGAAAAGGTCGCAACTCTTCTGTATGAGGTGATGCAAGACAATCCGGAGATATCGAAATTGTACTTAACTGGAGTGTTTTATTTTATGCTCTTGTACACGGGATCCAACCTGTTGCCTATAGCAAGGTTCCTGCGGCTCACGCACATGAGGCAGGCCTTTAGAGCTGATCAGAGCACGTCCGACATCATGCAGCGTTCGATATTAGGTCAGCTGCTGCCGGAAGCAATGGTCTGCTATCTGGAGAATCACGGAGCAGAGAAGTTCGCTCAGATATTCCTGGGAGAATGGGACACTCCAGAAGCCATTTGGAACAATGAAATGAGACGGATGTTGATAATGAAAGTATCGGCGCACATCGGCGAGTTCACGCCGCGGCTGCGTGCGCACGTGGCGGCGCGCTACCCCTACCTCGCGATACCGGCGCTGCGCTACCCGCAGCTGGACGCCGAGCTGTTCTGCAACATGTTCTACCTCAGGCATCTGTGCGACAACACCAGGTTCCCAGATTGGCCGATACCTGATCCGGTGGGGCTCTTGAAGGGTGTTCTCGAGGCATGGCGGCGTGAAGTTGAGAAGAAGCCGGCCGCCATGAGTGCGACGCAAGCGCTGGGCGTGCTGGGCCTGCCGCCCGACACCAGCGACGACGCCGTCGTGCGGAGGGCTTATTATCGACTGGCTCAGCAGCACCATCCAGATAAAAACCCTGAAGGAAGGGATCGTTTCGAGGCTGTAAATCAGGCGTACGAATTTCTGTGCAGCAGAAACGTGTGGACGGGAGACGGACCGAACACAAACAACATTGTACTTATATTGAGGACTCAAACTATACTTTTCCAAAGATATTCAGAAGTGTTGGCGCCGTACAAGTACGCGGGCTACCCGCAGCTGCTCCGCACGGCTCGGCTGGAGGCGGGCGCCGAGCAGCTGTTCGGTGCAGGCGGCGCCTTGCTGCCCGCCGCCTGCGAGCTCGCGCACGCCACGCTCTGCTGCTCCGCGCTCAACGCCGAGGAGCTGCGCCGCGAGCAAGGCCTAGAGGTTTTACAAGAGGCGCTGGCCCGGTGCGTGCCGCTGCTGGGCTCGGCGACGTCGGCGCAGGACCCGCTGGCGGCGGTGTGCGCACACTGCGCGCGCTGCTTCGCCGTGGCCGCCACGTTCCCGGCCTGCCGCGAGCGCTGCGCCCTCATGACGCAGCTCTGCATGGACATCGTGCCGCTACTCAAACGACCGAACCTGGGGGACACTGCGTGCGCGGGCGCGGAGGCGGCGGGCGCGCTGGCGGCGGACGCGCGGTGCCGGGCGCAGTTCGCGCGCGCCGGCCTGCTGCACGCGCTGCTGCCCGCCGCGCTGCAGTACGACTACACGCTCGCCGAGTCCGGCCTCGACACCGACGTCAAGGCCAACAAACAGGAGATATCGAATCAACTAGCGAAGAAGTGCGTGATAGCCTTGGCGGCGATGTACGGGCCGCACGAGGAGCCGGCGCCGGACGACGAGCGCATCCGCGACGCGCTGCACGTGCTGCTGACCCCGCACCTCTGCGGCAGACTCGCCGAGCCCGACCAGCACGAGCTCCTAAAGACTCTAACGTCGAACTGGCACACTCCGTACCTGGTGTGGGACAACAGCACGCGAGCCGAACTACAAGACGTGCTGCAGAACACCAGGGCCAACTACGAGGAGGAGGGGCCGCTGATAGCGCCCTTCACGTACTCGGCACACGCGGGCCTTCTGTCGGTCGGCGGAGTCTACATTGACATATACAACGAACAGCCTGACTTTCCTATTGAGAACCCACAACAGTTCATATTGGATCTGCTGAAATTCGTTCGGGAAGAAGTTAAAGGCGACATGACCTTGGAGCGAGTGGAGCACATAACACATGCTTTGACAGCACTCTCTAATGTCATTATTAAAAATCCAGGCGTCGAAATAAACTGCATCGGTCAGTTCGGTTTGATATTCGGCATGTTGTCGGGGCGCGGGCGGCCGCGACTACAGGACGCTGCGCTGCGGACGGTGCGCGCGGCGGCCGGCACGCGCGAGTGCGTGGACGACATCGCCGCCAGCTGCGTGCTCGGACACCTGCTCGCCGCGCTGCCAGAGCGCCCGACGCGCCCGGACGCGCTGCCCGCGCTCGCCGCGCTGCTCGCCAACACCGCGCTCGTCAAGGAGGCGCTCAACAAAGGTGCCGTGATATACCTACTGGATCTGTTCTGCAACAGTAAAGCGCCGGAGACGCGCGAAGCCGCCGTCGAACTCTTAGCGAGGATGATGGCAGACAAATTACACGGACCTAAGGTCCGCTTGGCGATATCTCGGTACGTGCCGGGCGTGTTCGCGGAGGCGATGCGCGAGTCGGGCCCCGCCAGCGCCGTGCACATGTACGACGCCGCGCATGAGCACCCCGAGCTCGTGTGGAGCGATGACGTCAGGCAGCGACTCGCCGAACACATCGCGCTACTCAAAGACAGACATCAAGCGAGACAGGCCCTCGACCCGAACGCGACGTTCGAGGACCGCGAGGTGGCGGCGGGCGAGCAGGCCTGGGCGCCGCCCGGCGAGGTGCTGGTGGGCGGGGTGTACCTCAAGCTCTATCTGCAGCACCCCTCCTGGAACCTGCGCAGCCCCAAGCGGTTCCTGCAGGACCTGCTCGCGGAGACCGTCGCCGCCTTACAGAAGGACTCCTCACAGGGCAGCAGAGGGGAGCTGTGCGCGCAGGCGCTGGTGGTGCTGCTGCGGTCCCGGCCCGCGCTCAGCGAGGCCTGCGCGCAGCTGGGCGAGCTGCCGCGCCTCACCAACATGCTGCCCTCCTGCCCCCTGCACATCGTCCCGGTGCTGGCCGCCGTCGCAGACACGCAGGCGTGTGTGATGGCGCTGACCCAGACCGACATAATGATCGGCATGAAGACCGCCGTGAAGTCGTGTCCCGAGGTGGTGGGCTCCGCGTGCGAGACGCTCTCCTCCATCTTCAGTAGCAACGTCAACACGGACCGCCTCGTGCTGCAGGCACTGGAATGCTCTCTGGTGAGCTCGCTGCTGTCGCTGCTGGAGTCGCGCGTGGAGGGCGGGTGCGCGGCGCGGCTGGTGGCGGCGCTGCAGGGCATGGCGCGCTCGCCGGCGCACGGACAGCGGGTGCGGGCCGCGCTCGCCGCCTGCGCCGTCTGGCCGCAGTACTCCGCGCAGAGGCACGACCTCTTCATCGCGCCGCCCGACCGACACTCCATCGCCGCTCCGCACTCGGCCGGCTACCTGACGGCGCCGCCGCTCAACATCCCGGCGCAGCCGCCCCCCATCGACTGA
Protein
MIPLKDNQDVASFLVTKHSWKGKYKRVFSIGTHGITTYNPDRLEVTNKWLYADVVTIASAKHSNSTANHDFTLVMKKEKKVDSMRFSSEHKCLILTEAFKYRHLFAEKPKDVFRYQAYKHHWSGTRLPIVLEVGPCALEQLDPSTHTQLAFYPYCDIQGILPVRDVAGGFMLAVGGYSRLHLFSNAMDHQIIINKMLELANLTLSISIKVLSATITLDDYHDQRFGKYSGDQHQTSLSEFIVHKVNPARHMEPMRRTLCLSDTCILERDPQTYSVVCLRPLSDVFALIRDPEQPQKFSIEYMNGQTRTYLAGERDALLASLLDGVRSAGQRDVHVRSTYTPRGYRLGPLHHSVDEETESNHLRLFQNPVGMTRAEVIERFNANIGYSGLIYSVSQDRLFAENKEKLITGALTALVNSVSSSATLPPRELEAQFHALRRLCASKVGFAAFTALTGFREWVGASVVAALRRGSAACSHAALDAVCALMQPMHHEPDLRQEQLNKASMLSSAQFLDGLLAMWAEHVSAGTGALVVAAMLDFLTFALCVPYSETTDGKQFDQLLEMVSNKGRVIFKLFEHPSMAIVKGAGLVMRALIEEGAGAVGTRMQALALAEAALPRHLLTALYAPARLHHRHLARHLVSLWLVDHAPAHNLLSRIMPSGLLGFLESAESPPAGALEEQRDEGRDNLQLAQAAHRPRASHWQALDRQVKYVEKHLEHYASLALQHWAQRTGRAGAGAGGAAREAVRERPVLLRRRRERVKSAANWPLFYYQFHRDHALPNLIWNHTTREELRNALENELRAFTSDREVAGNTLTSWNHAELELQYQCLQNEVKIGDYYLRILLEQKDNDDSPIRKSYEFFNDLYHRFLSTPKVEMKCMCLQAMSIVYGRYFEDIGPFADTRYIVQMLDRTCDRMERDRLVQFLSKLILHKRNVSEILEWNGIRILVELMTLAHLHTSRALIPAQSNVIEGPAEPGGNDREWYYNLEQGDKVQRKGPISLQQLKDLYKSGEITNKTKCWANSMEGWRAVGGIPQLKWTLVARGAPVFDESALAATILDLLITCSRYYPSRDEEDAVIRPLPRIKRLLSEPACLAHVVQLLLTFDPILVEKVATLLYEVMQDNPEISKLYLTGVFYFMLLYTGSNLLPIARFLRLTHMRQAFRADQSTSDIMQRSILGQLLPEAMVCYLENHGAEKFAQIFLGEWDTPEAIWNNEMRRMLIMKVSAHIGEFTPRLRAHVAARYPYLAIPALRYPQLDAELFCNMFYLRHLCDNTRFPDWPIPDPVGLLKGVLEAWRREVEKKPAAMSATQALGVLGLPPDTSDDAVVRRAYYRLAQQHHPDKNPEGRDRFEAVNQAYEFLCSRNVWTGDGPNTNNIVLILRTQTILFQRYSEVLAPYKYAGYPQLLRTARLEAGAEQLFGAGGALLPAACELAHATLCCSALNAEELRREQGLEVLQEALARCVPLLGSATSAQDPLAAVCAHCARCFAVAATFPACRERCALMTQLCMDIVPLLKRPNLGDTACAGAEAAGALAADARCRAQFARAGLLHALLPAALQYDYTLAESGLDTDVKANKQEISNQLAKKCVIALAAMYGPHEEPAPDDERIRDALHVLLTPHLCGRLAEPDQHELLKTLTSNWHTPYLVWDNSTRAELQDVLQNTRANYEEEGPLIAPFTYSAHAGLLSVGGVYIDIYNEQPDFPIENPQQFILDLLKFVREEVKGDMTLERVEHITHALTALSNVIIKNPGVEINCIGQFGLIFGMLSGRGRPRLQDAALRTVRAAAGTRECVDDIAASCVLGHLLAALPERPTRPDALPALAALLANTALVKEALNKGAVIYLLDLFCNSKAPETREAAVELLARMMADKLHGPKVRLAISRYVPGVFAEAMRESGPASAVHMYDAAHEHPELVWSDDVRQRLAEHIALLKDRHQARQALDPNATFEDREVAAGEQAWAPPGEVLVGGVYLKLYLQHPSWNLRSPKRFLQDLLAETVAALQKDSSQGSRGELCAQALVVLLRSRPALSEACAQLGELPRLTNMLPSCPLHIVPVLAAVADTQACVMALTQTDIMIGMKTAVKSCPEVVGSACETLSSIFSSNVNTDRLVLQALECSLVSSLLSLLESRVEGGCAARLVAALQGMARSPAHGQRVRAALAACAVWPQYSAQRHDLFIAPPDRHSIAAPHSAGYLTAPPLNIPAQPPPID
Summary
Similarity
Belongs to the histidine acid phosphatase family.
Uniprot
A0A194QIE8
D6WQC2
A0A088AL57
A0A2A3ERU6
A0A154P3S9
A0A1W4WLN4
+ More
A0A0C9RF69 E2C591 A0A3L8DNN9 A0A026WRT1 A0A232EW23 F4X7Z1 A0A0J7P3Q6 E1ZXT6 A0A1Y1L7D0 A0A0M9A3Z2 A0A151I1S1 A0A195FXD4 U4TXZ4 A0A1W4WLV8 A0A151IDQ8 A0A067RE14 N6TIY1 A0A0L7QTS4 A0A1Y1LB56 A0A151IRV5 A0A158NWG3 A0A151WGD4 A0A2J7R5G3 A0A0T6AT64 A0A2R2MTA1 A0A2C9K7Q6 A0A210Q622 A0A1U7RMC0 A0A2C9K7K9 A0A3Q0GCZ4 A0A3S1B5Q9 A0A151P2Z2 A0A0L8HG32 A0A287AXH6 A0A286ZQV5 A0A1D1VI44 A0A1S3WUH7 F1MDR7 G1K9A0 F1SPE9 A0A2U4B924 A0A2U4B983 A0A341CRJ8 A0A3Q7Q7D6 A0A341CUC6 A0A2U3X1M8 A0A2U3ZWK8 K1RBJ3 U3KDB1 K9IPK5 A0A0L8HGH6 A0A093QZR7 A0A087QIS0 A0A091SUK3 G1NG41 A0A091J823 K9IPG9 E2R6R6 A0A3Q7T2T1 A0A3Q7TND4 A0A093RMV2 A0A091LYI3 M3W143 A0A384DIJ0 A0A337S082 A0A3Q7UVT7 A0A3Q7VKW5 A0A1D5NZ71 A0A093PHU3 A0A2I2UIN5 S7MPA1 A0A087VNZ9 A0A091QMR0 A0A091JYG3 A0A3P4S5G6 A0A1L8FWJ0 A0A091NDZ0 W5K4K3 A0A093IG07 A0A2I0MRJ2 A0A091FMI7 A0A3L8SYB6 K7FX46 H0Z1S7 K7FX58 A0A2T7PZ08 G1SDM6 A0A091V0R1 A0A093KCH5 A0A3P8NR98 A0A2I2UK41 A0A3P9CS98 A0A093IYR8
A0A0C9RF69 E2C591 A0A3L8DNN9 A0A026WRT1 A0A232EW23 F4X7Z1 A0A0J7P3Q6 E1ZXT6 A0A1Y1L7D0 A0A0M9A3Z2 A0A151I1S1 A0A195FXD4 U4TXZ4 A0A1W4WLV8 A0A151IDQ8 A0A067RE14 N6TIY1 A0A0L7QTS4 A0A1Y1LB56 A0A151IRV5 A0A158NWG3 A0A151WGD4 A0A2J7R5G3 A0A0T6AT64 A0A2R2MTA1 A0A2C9K7Q6 A0A210Q622 A0A1U7RMC0 A0A2C9K7K9 A0A3Q0GCZ4 A0A3S1B5Q9 A0A151P2Z2 A0A0L8HG32 A0A287AXH6 A0A286ZQV5 A0A1D1VI44 A0A1S3WUH7 F1MDR7 G1K9A0 F1SPE9 A0A2U4B924 A0A2U4B983 A0A341CRJ8 A0A3Q7Q7D6 A0A341CUC6 A0A2U3X1M8 A0A2U3ZWK8 K1RBJ3 U3KDB1 K9IPK5 A0A0L8HGH6 A0A093QZR7 A0A087QIS0 A0A091SUK3 G1NG41 A0A091J823 K9IPG9 E2R6R6 A0A3Q7T2T1 A0A3Q7TND4 A0A093RMV2 A0A091LYI3 M3W143 A0A384DIJ0 A0A337S082 A0A3Q7UVT7 A0A3Q7VKW5 A0A1D5NZ71 A0A093PHU3 A0A2I2UIN5 S7MPA1 A0A087VNZ9 A0A091QMR0 A0A091JYG3 A0A3P4S5G6 A0A1L8FWJ0 A0A091NDZ0 W5K4K3 A0A093IG07 A0A2I0MRJ2 A0A091FMI7 A0A3L8SYB6 K7FX46 H0Z1S7 K7FX58 A0A2T7PZ08 G1SDM6 A0A091V0R1 A0A093KCH5 A0A3P8NR98 A0A2I2UK41 A0A3P9CS98 A0A093IYR8
Pubmed
EMBL
KQ459185
KPJ03226.1
KQ971354
EFA06092.2
KZ288191
PBC34458.1
+ More
KQ434809 KZC06599.1 GBYB01015224 JAG84991.1 GL452767 EFN76912.1 QOIP01000006 RLU21843.1 KK107128 EZA58396.1 NNAY01001930 OXU22526.1 GL888900 EGI57408.1 LBMM01000182 KMR04574.1 GL435089 EFN74007.1 GEZM01065931 JAV68290.1 KQ435741 KOX76760.1 KQ976554 KYM80799.1 KQ981193 KYN45086.1 KB631740 ERL85687.1 KQ977907 KYM98836.1 KK852527 KDR22007.1 APGK01024435 KB740546 ENN80399.1 KQ414740 KOC62067.1 GEZM01065933 JAV68287.1 KQ981104 KYN09453.1 ADTU01027956 KQ983178 KYQ46884.1 NEVH01006994 PNF36068.1 LJIG01022870 KRT78303.1 NEDP02004861 OWF44178.1 RQTK01000726 RUS75597.1 AKHW03001210 KYO43350.1 KQ418215 KOF88208.1 AEMK02000088 DQIR01222424 DQIR01280933 HDB77901.1 DQIR01222423 HDB77900.1 BDGG01000004 GAU98158.1 JH817258 EKC31461.1 AGTO01001245 GABZ01003417 JAA50108.1 KOF88209.1 KL432050 KFW91885.1 KL225640 KFM01124.1 KK487238 KFQ62349.1 KK501715 KFP17149.1 GABZ01003420 JAA50105.1 AAEX03013595 AAEX03013596 KL225452 KFW72199.1 KK511846 KFP63384.1 AANG04001816 AADN05000034 KL669380 KFW75991.1 KE161960 EPQ06114.1 KL500368 KFO14341.1 KK678250 KFQ10744.1 KK534939 KFP29146.1 CYRY02046697 VCX42453.1 CM004476 OCT75945.1 KK845621 KFP87292.1 KL215597 KFV65686.1 AKCR02000004 PKK32297.1 KL447199 KFO70803.1 QUSF01000003 RLW11573.1 AGCU01153347 AGCU01153348 AGCU01153349 AGCU01153350 AGCU01153351 AGCU01153352 AGCU01153353 AGCU01153354 ABQF01014118 PZQS01000001 PVD38649.1 KL410402 KFQ96521.1 KL206987 KFV87974.1 KK571764 KFW07885.1
KQ434809 KZC06599.1 GBYB01015224 JAG84991.1 GL452767 EFN76912.1 QOIP01000006 RLU21843.1 KK107128 EZA58396.1 NNAY01001930 OXU22526.1 GL888900 EGI57408.1 LBMM01000182 KMR04574.1 GL435089 EFN74007.1 GEZM01065931 JAV68290.1 KQ435741 KOX76760.1 KQ976554 KYM80799.1 KQ981193 KYN45086.1 KB631740 ERL85687.1 KQ977907 KYM98836.1 KK852527 KDR22007.1 APGK01024435 KB740546 ENN80399.1 KQ414740 KOC62067.1 GEZM01065933 JAV68287.1 KQ981104 KYN09453.1 ADTU01027956 KQ983178 KYQ46884.1 NEVH01006994 PNF36068.1 LJIG01022870 KRT78303.1 NEDP02004861 OWF44178.1 RQTK01000726 RUS75597.1 AKHW03001210 KYO43350.1 KQ418215 KOF88208.1 AEMK02000088 DQIR01222424 DQIR01280933 HDB77901.1 DQIR01222423 HDB77900.1 BDGG01000004 GAU98158.1 JH817258 EKC31461.1 AGTO01001245 GABZ01003417 JAA50108.1 KOF88209.1 KL432050 KFW91885.1 KL225640 KFM01124.1 KK487238 KFQ62349.1 KK501715 KFP17149.1 GABZ01003420 JAA50105.1 AAEX03013595 AAEX03013596 KL225452 KFW72199.1 KK511846 KFP63384.1 AANG04001816 AADN05000034 KL669380 KFW75991.1 KE161960 EPQ06114.1 KL500368 KFO14341.1 KK678250 KFQ10744.1 KK534939 KFP29146.1 CYRY02046697 VCX42453.1 CM004476 OCT75945.1 KK845621 KFP87292.1 KL215597 KFV65686.1 AKCR02000004 PKK32297.1 KL447199 KFO70803.1 QUSF01000003 RLW11573.1 AGCU01153347 AGCU01153348 AGCU01153349 AGCU01153350 AGCU01153351 AGCU01153352 AGCU01153353 AGCU01153354 ABQF01014118 PZQS01000001 PVD38649.1 KL410402 KFQ96521.1 KL206987 KFV87974.1 KK571764 KFW07885.1
Proteomes
UP000053268
UP000007266
UP000005203
UP000242457
UP000076502
UP000192223
+ More
UP000008237 UP000279307 UP000053097 UP000215335 UP000007755 UP000036403 UP000000311 UP000053105 UP000078540 UP000078541 UP000030742 UP000078542 UP000027135 UP000019118 UP000053825 UP000078492 UP000005205 UP000075809 UP000235965 UP000085678 UP000076420 UP000242188 UP000189705 UP000271974 UP000050525 UP000053454 UP000008227 UP000186922 UP000079721 UP000009136 UP000001646 UP000245320 UP000252040 UP000286641 UP000245340 UP000005408 UP000016665 UP000053286 UP000001645 UP000053119 UP000002254 UP000286640 UP000054081 UP000011712 UP000261680 UP000286642 UP000000539 UP000053258 UP000186698 UP000018467 UP000053875 UP000053872 UP000053760 UP000276834 UP000007267 UP000007754 UP000245119 UP000001811 UP000053283 UP000053584 UP000265100 UP000265160
UP000008237 UP000279307 UP000053097 UP000215335 UP000007755 UP000036403 UP000000311 UP000053105 UP000078540 UP000078541 UP000030742 UP000078542 UP000027135 UP000019118 UP000053825 UP000078492 UP000005205 UP000075809 UP000235965 UP000085678 UP000076420 UP000242188 UP000189705 UP000271974 UP000050525 UP000053454 UP000008227 UP000186922 UP000079721 UP000009136 UP000001646 UP000245320 UP000252040 UP000286641 UP000245340 UP000005408 UP000016665 UP000053286 UP000001645 UP000053119 UP000002254 UP000286640 UP000054081 UP000011712 UP000261680 UP000286642 UP000000539 UP000053258 UP000186698 UP000018467 UP000053875 UP000053872 UP000053760 UP000276834 UP000007267 UP000007754 UP000245119 UP000001811 UP000053283 UP000053584 UP000265100 UP000265160
PRIDE
Interpro
IPR025640
GYF_2
+ More
IPR001623 DnaJ_domain
IPR036869 J_dom_sf
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR035445 GYF-like_dom_sf
IPR029033 His_PPase_superfam
IPR000560 His_Pase_clade-2
IPR033379 Acid_Pase_AS
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR001623 DnaJ_domain
IPR036869 J_dom_sf
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR035445 GYF-like_dom_sf
IPR029033 His_PPase_superfam
IPR000560 His_Pase_clade-2
IPR033379 Acid_Pase_AS
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
SUPFAM
ProteinModelPortal
A0A194QIE8
D6WQC2
A0A088AL57
A0A2A3ERU6
A0A154P3S9
A0A1W4WLN4
+ More
A0A0C9RF69 E2C591 A0A3L8DNN9 A0A026WRT1 A0A232EW23 F4X7Z1 A0A0J7P3Q6 E1ZXT6 A0A1Y1L7D0 A0A0M9A3Z2 A0A151I1S1 A0A195FXD4 U4TXZ4 A0A1W4WLV8 A0A151IDQ8 A0A067RE14 N6TIY1 A0A0L7QTS4 A0A1Y1LB56 A0A151IRV5 A0A158NWG3 A0A151WGD4 A0A2J7R5G3 A0A0T6AT64 A0A2R2MTA1 A0A2C9K7Q6 A0A210Q622 A0A1U7RMC0 A0A2C9K7K9 A0A3Q0GCZ4 A0A3S1B5Q9 A0A151P2Z2 A0A0L8HG32 A0A287AXH6 A0A286ZQV5 A0A1D1VI44 A0A1S3WUH7 F1MDR7 G1K9A0 F1SPE9 A0A2U4B924 A0A2U4B983 A0A341CRJ8 A0A3Q7Q7D6 A0A341CUC6 A0A2U3X1M8 A0A2U3ZWK8 K1RBJ3 U3KDB1 K9IPK5 A0A0L8HGH6 A0A093QZR7 A0A087QIS0 A0A091SUK3 G1NG41 A0A091J823 K9IPG9 E2R6R6 A0A3Q7T2T1 A0A3Q7TND4 A0A093RMV2 A0A091LYI3 M3W143 A0A384DIJ0 A0A337S082 A0A3Q7UVT7 A0A3Q7VKW5 A0A1D5NZ71 A0A093PHU3 A0A2I2UIN5 S7MPA1 A0A087VNZ9 A0A091QMR0 A0A091JYG3 A0A3P4S5G6 A0A1L8FWJ0 A0A091NDZ0 W5K4K3 A0A093IG07 A0A2I0MRJ2 A0A091FMI7 A0A3L8SYB6 K7FX46 H0Z1S7 K7FX58 A0A2T7PZ08 G1SDM6 A0A091V0R1 A0A093KCH5 A0A3P8NR98 A0A2I2UK41 A0A3P9CS98 A0A093IYR8
A0A0C9RF69 E2C591 A0A3L8DNN9 A0A026WRT1 A0A232EW23 F4X7Z1 A0A0J7P3Q6 E1ZXT6 A0A1Y1L7D0 A0A0M9A3Z2 A0A151I1S1 A0A195FXD4 U4TXZ4 A0A1W4WLV8 A0A151IDQ8 A0A067RE14 N6TIY1 A0A0L7QTS4 A0A1Y1LB56 A0A151IRV5 A0A158NWG3 A0A151WGD4 A0A2J7R5G3 A0A0T6AT64 A0A2R2MTA1 A0A2C9K7Q6 A0A210Q622 A0A1U7RMC0 A0A2C9K7K9 A0A3Q0GCZ4 A0A3S1B5Q9 A0A151P2Z2 A0A0L8HG32 A0A287AXH6 A0A286ZQV5 A0A1D1VI44 A0A1S3WUH7 F1MDR7 G1K9A0 F1SPE9 A0A2U4B924 A0A2U4B983 A0A341CRJ8 A0A3Q7Q7D6 A0A341CUC6 A0A2U3X1M8 A0A2U3ZWK8 K1RBJ3 U3KDB1 K9IPK5 A0A0L8HGH6 A0A093QZR7 A0A087QIS0 A0A091SUK3 G1NG41 A0A091J823 K9IPG9 E2R6R6 A0A3Q7T2T1 A0A3Q7TND4 A0A093RMV2 A0A091LYI3 M3W143 A0A384DIJ0 A0A337S082 A0A3Q7UVT7 A0A3Q7VKW5 A0A1D5NZ71 A0A093PHU3 A0A2I2UIN5 S7MPA1 A0A087VNZ9 A0A091QMR0 A0A091JYG3 A0A3P4S5G6 A0A1L8FWJ0 A0A091NDZ0 W5K4K3 A0A093IG07 A0A2I0MRJ2 A0A091FMI7 A0A3L8SYB6 K7FX46 H0Z1S7 K7FX58 A0A2T7PZ08 G1SDM6 A0A091V0R1 A0A093KCH5 A0A3P8NR98 A0A2I2UK41 A0A3P9CS98 A0A093IYR8
PDB
2OCH
E-value=6.46736e-07,
Score=135
Ontologies
KEGG
GO
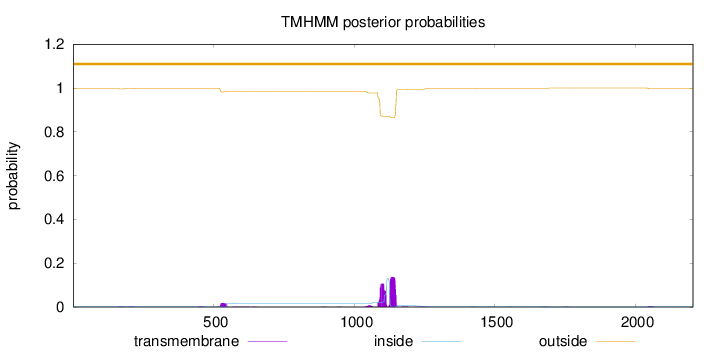
Topology
Length:
2205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.32622999999996
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.00345
outside
1 - 2205
Population Genetic Test Statistics
Pi
233.498627
Theta
172.371477
Tajima's D
1.258014
CLR
0.309095
CSRT
0.726563671816409
Interpretation
Uncertain
