Gene
KWMTBOMO11674
Pre Gene Modal
BGIBMGA003980
Annotation
PREDICTED:_GTP-binding_protein_Rit2?_partial_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.396 Mitochondrial Reliability : 1.304 Nuclear Reliability : 1.079
Sequence
CDS
ATGGCGGGCGAGGCTGCGGGCACGTCGCGAGGGTTGCGCGTATACAAGATCGTGGTACTGGGCGACGGCGGGGTCGGCAAGTCTGCGGTCACACTCCAGTTCGTCAGCCACAGTTTTTTGGACTACCATGACCCAACCATAGAGGACTCCTATCAACAACAAGCAGTAATTGATGGAGAACCAGCACTGCTGGACATTCTGGATACAGCTGGTCAGGTGGAGTTCACTGCGATGCGGGAGCAGTACATGCGTTGCGGGGAAGGCTTTGTGCTGTGCTACTCGGTGACGGACCGGCGATCGTTCCGCGCTGCCGCCGAGTACAGGAGGCTGCTGGCGCAGGCGCGTCCCTCCGAGAGGCTCCCACTCGTGCTGGTTGGAAACAAACTTGACCTAGCTCCGAGACTTAGACAGGTAACGACCGAGGAAGGCAAGGAGCTGGCCACACAGCTGGGGTGTCCGTTCTACGAGACTTCGGCGGCGCTAAGGCACTTCGTCGATGACGCATTCCACGCACTCGTCCGAGAGATACGGCGCCTAGAGAGACAGAGACAGAAAAACTATCATAACAAAAGAGATATTTGA
Protein
MAGEAAGTSRGLRVYKIVVLGDGGVGKSAVTLQFVSHSFLDYHDPTIEDSYQQQAVIDGEPALLDILDTAGQVEFTAMREQYMRCGEGFVLCYSVTDRRSFRAAAEYRRLLAQARPSERLPLVLVGNKLDLAPRLRQVTTEEGKELATQLGCPFYETSAALRHFVDDAFHALVREIRRLERQRQKNYHNKRDI
Summary
Uniprot
A0A194QYD0
A0A194QIF5
A0A2H1WUQ0
A0A2A4JPR2
A0A212FP27
A0A212FAS3
+ More
A0A182QIY1 A0A182MV23 A0A2Y9D0Y8 A0A182UWY0 A0A182TXD8 A0A182LBH2 Q7Q9M1 Q16WL0 A0A1S4FLW5 A0A2M4AX49 W5JRD3 A0A182FED7 A0A182IL66 A0A1W4WJF0 A0A154P2J3 A0A084W2E6 B0X442 A0A0M9A0C0 Q28WT4 B4H6A6 A0A0M4EUV1 A0A0Q9WWT8 A0A1Y1M7R4 A0A088ALI8 A0A182XJC9 D6WSK4 A0A0L7QRQ4 A0A182R3D2 A0A3B0J020 B4KQB0 A0A1W4UZR2 A0A182N857 B3MF50 B4LL57 B4J9Y2 A0A182WAE2 B4P6R4 B3NPX5 E9JAW2 H9J393 A0A336MUA4 Q7JMZ0 B4HSJ7 B4QHF1 A0A1B0CCY4 E2BBL8 A0A023GIU9 A0A1J1JAH2 A0A0L0CB96 E2AT49 T1PID6 K7J0D5 A0A026W802 A0A232FLD9 F4WHH5 A0A151WNM9 A0A3L8DTI3 A0A195FTZ3 A0A182YIH8 A0A151J7Q3 A0A2J7Q6A4 A0A195BUW7 A0A158N926 A0A0A1XBH7 A0A034VNN2 A0A0A1X7F4 A0A0P4WE30 A0A0P4WKH4 A0A067QYA8 A0A0K8UW67 W8B4B5 A0A1I8NRK7 A0A182PCB9 A0A1E1X3C1 A0A1E1XTE3 A0A293N2T1 G3MLW8 A0A224YZJ1 A0A131YH10 L7M7Q2 V5I4R3 A0A131XC14 A0A2R5LJJ7 L7LTJ3 Q16EF0 A0A2M4C168 A0A182SAP5 T1IWJ0 A0A2B4S198 E0VQH6 E5L9R9 A0A3M6T989 A0A2P2I560 G1KPH1
A0A182QIY1 A0A182MV23 A0A2Y9D0Y8 A0A182UWY0 A0A182TXD8 A0A182LBH2 Q7Q9M1 Q16WL0 A0A1S4FLW5 A0A2M4AX49 W5JRD3 A0A182FED7 A0A182IL66 A0A1W4WJF0 A0A154P2J3 A0A084W2E6 B0X442 A0A0M9A0C0 Q28WT4 B4H6A6 A0A0M4EUV1 A0A0Q9WWT8 A0A1Y1M7R4 A0A088ALI8 A0A182XJC9 D6WSK4 A0A0L7QRQ4 A0A182R3D2 A0A3B0J020 B4KQB0 A0A1W4UZR2 A0A182N857 B3MF50 B4LL57 B4J9Y2 A0A182WAE2 B4P6R4 B3NPX5 E9JAW2 H9J393 A0A336MUA4 Q7JMZ0 B4HSJ7 B4QHF1 A0A1B0CCY4 E2BBL8 A0A023GIU9 A0A1J1JAH2 A0A0L0CB96 E2AT49 T1PID6 K7J0D5 A0A026W802 A0A232FLD9 F4WHH5 A0A151WNM9 A0A3L8DTI3 A0A195FTZ3 A0A182YIH8 A0A151J7Q3 A0A2J7Q6A4 A0A195BUW7 A0A158N926 A0A0A1XBH7 A0A034VNN2 A0A0A1X7F4 A0A0P4WE30 A0A0P4WKH4 A0A067QYA8 A0A0K8UW67 W8B4B5 A0A1I8NRK7 A0A182PCB9 A0A1E1X3C1 A0A1E1XTE3 A0A293N2T1 G3MLW8 A0A224YZJ1 A0A131YH10 L7M7Q2 V5I4R3 A0A131XC14 A0A2R5LJJ7 L7LTJ3 Q16EF0 A0A2M4C168 A0A182SAP5 T1IWJ0 A0A2B4S198 E0VQH6 E5L9R9 A0A3M6T989 A0A2P2I560 G1KPH1
Pubmed
26354079
22118469
20966253
12364791
14747013
17210077
+ More
17510324 20920257 23761445 24438588 15632085 17994087 28004739 18362917 19820115 17550304 21282665 19121390 8918462 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20798317 26108605 25315136 20075255 24508170 28648823 21719571 30249741 25244985 21347285 25830018 25348373 24845553 24495485 28503490 29209593 22216098 28797301 26830274 25576852 25765539 28049606 20566863 20978910 30382153
17510324 20920257 23761445 24438588 15632085 17994087 28004739 18362917 19820115 17550304 21282665 19121390 8918462 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20798317 26108605 25315136 20075255 24508170 28648823 21719571 30249741 25244985 21347285 25830018 25348373 24845553 24495485 28503490 29209593 22216098 28797301 26830274 25576852 25765539 28049606 20566863 20978910 30382153
EMBL
KQ460949
KPJ10472.1
KQ459185
KPJ03231.1
ODYU01011205
SOQ56791.1
+ More
NWSH01000825 PCG73991.1 AGBW02003976 OWR55485.1 AGBW02009432 OWR50830.1 AXCN02001001 AXCM01003731 APCN01002896 AAAB01008900 EAA09461.4 CH477562 EAT38979.1 GGFK01012043 MBW45364.1 ADMH02000689 ETN65309.1 KQ434807 KZC06159.1 ATLV01019609 KE525275 KFB44390.1 DS232328 EDS40119.1 KQ435794 KOX73746.1 CM000071 EAL26582.1 CH479213 EDW33330.1 CP012524 ALC41295.1 CH964282 KRG00245.1 GEZM01042297 JAV79956.1 KQ971354 EFA05904.1 KQ414775 KOC61308.1 OUUW01000001 SPP73955.1 CH933808 EDW09238.1 CH902619 EDV37678.1 CH940648 EDW61864.2 CH916367 EDW02569.1 CM000158 EDW91991.1 CH954179 EDV55822.1 GL770679 EFZ10020.1 BABH01021727 BABH01021728 UFQS01002634 UFQT01002634 SSX14416.1 SSX33826.1 AE013599 Y07564 AAF58065.1 ACZ94437.1 CAA68849.1 CH480816 EDW48075.1 CM000362 CM002911 EDX07296.1 KMY94159.1 AJWK01007219 GL447128 EFN86895.1 GBBM01002718 JAC32700.1 CVRI01000075 CRL08558.1 JRES01000655 KNC29507.1 GL442495 EFN63389.1 KA648562 AFP63191.1 KK107372 EZA51776.1 NNAY01000081 OXU31157.1 GL888161 EGI66329.1 KQ982907 KYQ49474.1 QOIP01000004 RLU23694.1 KQ981268 KYN43928.1 KQ979640 KYN20039.1 NEVH01017534 PNF24111.1 KQ976405 KYM91875.1 ADTU01009051 GBXI01017119 GBXI01006000 JAC97172.1 JAD08292.1 GAKP01015794 GAKP01015792 GAKP01015789 JAC43158.1 GBXI01007679 JAD06613.1 GDRN01065397 JAI64692.1 GDRN01065398 JAI64691.1 KK853111 KDR11212.1 GDHF01021385 JAI30929.1 GAMC01013133 JAB93422.1 GFAC01005444 JAT93744.1 GFAA01001155 JAU02280.1 GFWV01022369 MAA47096.1 JO842869 AEO34486.1 GFPF01011209 MAA22355.1 GEDV01010028 JAP78529.1 GACK01005851 JAA59183.1 GANP01001040 JAB83428.1 GEFH01004946 JAP63635.1 GGLE01005596 MBY09722.1 GACK01010107 JAA54927.1 CH478791 EAT32608.1 GGFJ01009597 MBW58738.1 JH431620 LSMT01000235 PFX22570.1 DS235430 EEB15632.1 HQ384050 ADQ64543.1 RCHS01004063 RMX37976.1 IACF01003521 LAB69135.1
NWSH01000825 PCG73991.1 AGBW02003976 OWR55485.1 AGBW02009432 OWR50830.1 AXCN02001001 AXCM01003731 APCN01002896 AAAB01008900 EAA09461.4 CH477562 EAT38979.1 GGFK01012043 MBW45364.1 ADMH02000689 ETN65309.1 KQ434807 KZC06159.1 ATLV01019609 KE525275 KFB44390.1 DS232328 EDS40119.1 KQ435794 KOX73746.1 CM000071 EAL26582.1 CH479213 EDW33330.1 CP012524 ALC41295.1 CH964282 KRG00245.1 GEZM01042297 JAV79956.1 KQ971354 EFA05904.1 KQ414775 KOC61308.1 OUUW01000001 SPP73955.1 CH933808 EDW09238.1 CH902619 EDV37678.1 CH940648 EDW61864.2 CH916367 EDW02569.1 CM000158 EDW91991.1 CH954179 EDV55822.1 GL770679 EFZ10020.1 BABH01021727 BABH01021728 UFQS01002634 UFQT01002634 SSX14416.1 SSX33826.1 AE013599 Y07564 AAF58065.1 ACZ94437.1 CAA68849.1 CH480816 EDW48075.1 CM000362 CM002911 EDX07296.1 KMY94159.1 AJWK01007219 GL447128 EFN86895.1 GBBM01002718 JAC32700.1 CVRI01000075 CRL08558.1 JRES01000655 KNC29507.1 GL442495 EFN63389.1 KA648562 AFP63191.1 KK107372 EZA51776.1 NNAY01000081 OXU31157.1 GL888161 EGI66329.1 KQ982907 KYQ49474.1 QOIP01000004 RLU23694.1 KQ981268 KYN43928.1 KQ979640 KYN20039.1 NEVH01017534 PNF24111.1 KQ976405 KYM91875.1 ADTU01009051 GBXI01017119 GBXI01006000 JAC97172.1 JAD08292.1 GAKP01015794 GAKP01015792 GAKP01015789 JAC43158.1 GBXI01007679 JAD06613.1 GDRN01065397 JAI64692.1 GDRN01065398 JAI64691.1 KK853111 KDR11212.1 GDHF01021385 JAI30929.1 GAMC01013133 JAB93422.1 GFAC01005444 JAT93744.1 GFAA01001155 JAU02280.1 GFWV01022369 MAA47096.1 JO842869 AEO34486.1 GFPF01011209 MAA22355.1 GEDV01010028 JAP78529.1 GACK01005851 JAA59183.1 GANP01001040 JAB83428.1 GEFH01004946 JAP63635.1 GGLE01005596 MBY09722.1 GACK01010107 JAA54927.1 CH478791 EAT32608.1 GGFJ01009597 MBW58738.1 JH431620 LSMT01000235 PFX22570.1 DS235430 EEB15632.1 HQ384050 ADQ64543.1 RCHS01004063 RMX37976.1 IACF01003521 LAB69135.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000075886
UP000075883
+ More
UP000075840 UP000075903 UP000075902 UP000075882 UP000007062 UP000008820 UP000000673 UP000069272 UP000075880 UP000192223 UP000076502 UP000030765 UP000002320 UP000053105 UP000001819 UP000008744 UP000092553 UP000007798 UP000005203 UP000076407 UP000007266 UP000053825 UP000075900 UP000268350 UP000009192 UP000192221 UP000075884 UP000007801 UP000008792 UP000001070 UP000075920 UP000002282 UP000008711 UP000005204 UP000000803 UP000001292 UP000000304 UP000092461 UP000008237 UP000183832 UP000037069 UP000000311 UP000095301 UP000002358 UP000053097 UP000215335 UP000007755 UP000075809 UP000279307 UP000078541 UP000076408 UP000078492 UP000235965 UP000078540 UP000005205 UP000027135 UP000095300 UP000075885 UP000075901 UP000225706 UP000009046 UP000275408 UP000001646
UP000075840 UP000075903 UP000075902 UP000075882 UP000007062 UP000008820 UP000000673 UP000069272 UP000075880 UP000192223 UP000076502 UP000030765 UP000002320 UP000053105 UP000001819 UP000008744 UP000092553 UP000007798 UP000005203 UP000076407 UP000007266 UP000053825 UP000075900 UP000268350 UP000009192 UP000192221 UP000075884 UP000007801 UP000008792 UP000001070 UP000075920 UP000002282 UP000008711 UP000005204 UP000000803 UP000001292 UP000000304 UP000092461 UP000008237 UP000183832 UP000037069 UP000000311 UP000095301 UP000002358 UP000053097 UP000215335 UP000007755 UP000075809 UP000279307 UP000078541 UP000076408 UP000078492 UP000235965 UP000078540 UP000005205 UP000027135 UP000095300 UP000075885 UP000075901 UP000225706 UP000009046 UP000275408 UP000001646
Interpro
ProteinModelPortal
A0A194QYD0
A0A194QIF5
A0A2H1WUQ0
A0A2A4JPR2
A0A212FP27
A0A212FAS3
+ More
A0A182QIY1 A0A182MV23 A0A2Y9D0Y8 A0A182UWY0 A0A182TXD8 A0A182LBH2 Q7Q9M1 Q16WL0 A0A1S4FLW5 A0A2M4AX49 W5JRD3 A0A182FED7 A0A182IL66 A0A1W4WJF0 A0A154P2J3 A0A084W2E6 B0X442 A0A0M9A0C0 Q28WT4 B4H6A6 A0A0M4EUV1 A0A0Q9WWT8 A0A1Y1M7R4 A0A088ALI8 A0A182XJC9 D6WSK4 A0A0L7QRQ4 A0A182R3D2 A0A3B0J020 B4KQB0 A0A1W4UZR2 A0A182N857 B3MF50 B4LL57 B4J9Y2 A0A182WAE2 B4P6R4 B3NPX5 E9JAW2 H9J393 A0A336MUA4 Q7JMZ0 B4HSJ7 B4QHF1 A0A1B0CCY4 E2BBL8 A0A023GIU9 A0A1J1JAH2 A0A0L0CB96 E2AT49 T1PID6 K7J0D5 A0A026W802 A0A232FLD9 F4WHH5 A0A151WNM9 A0A3L8DTI3 A0A195FTZ3 A0A182YIH8 A0A151J7Q3 A0A2J7Q6A4 A0A195BUW7 A0A158N926 A0A0A1XBH7 A0A034VNN2 A0A0A1X7F4 A0A0P4WE30 A0A0P4WKH4 A0A067QYA8 A0A0K8UW67 W8B4B5 A0A1I8NRK7 A0A182PCB9 A0A1E1X3C1 A0A1E1XTE3 A0A293N2T1 G3MLW8 A0A224YZJ1 A0A131YH10 L7M7Q2 V5I4R3 A0A131XC14 A0A2R5LJJ7 L7LTJ3 Q16EF0 A0A2M4C168 A0A182SAP5 T1IWJ0 A0A2B4S198 E0VQH6 E5L9R9 A0A3M6T989 A0A2P2I560 G1KPH1
A0A182QIY1 A0A182MV23 A0A2Y9D0Y8 A0A182UWY0 A0A182TXD8 A0A182LBH2 Q7Q9M1 Q16WL0 A0A1S4FLW5 A0A2M4AX49 W5JRD3 A0A182FED7 A0A182IL66 A0A1W4WJF0 A0A154P2J3 A0A084W2E6 B0X442 A0A0M9A0C0 Q28WT4 B4H6A6 A0A0M4EUV1 A0A0Q9WWT8 A0A1Y1M7R4 A0A088ALI8 A0A182XJC9 D6WSK4 A0A0L7QRQ4 A0A182R3D2 A0A3B0J020 B4KQB0 A0A1W4UZR2 A0A182N857 B3MF50 B4LL57 B4J9Y2 A0A182WAE2 B4P6R4 B3NPX5 E9JAW2 H9J393 A0A336MUA4 Q7JMZ0 B4HSJ7 B4QHF1 A0A1B0CCY4 E2BBL8 A0A023GIU9 A0A1J1JAH2 A0A0L0CB96 E2AT49 T1PID6 K7J0D5 A0A026W802 A0A232FLD9 F4WHH5 A0A151WNM9 A0A3L8DTI3 A0A195FTZ3 A0A182YIH8 A0A151J7Q3 A0A2J7Q6A4 A0A195BUW7 A0A158N926 A0A0A1XBH7 A0A034VNN2 A0A0A1X7F4 A0A0P4WE30 A0A0P4WKH4 A0A067QYA8 A0A0K8UW67 W8B4B5 A0A1I8NRK7 A0A182PCB9 A0A1E1X3C1 A0A1E1XTE3 A0A293N2T1 G3MLW8 A0A224YZJ1 A0A131YH10 L7M7Q2 V5I4R3 A0A131XC14 A0A2R5LJJ7 L7LTJ3 Q16EF0 A0A2M4C168 A0A182SAP5 T1IWJ0 A0A2B4S198 E0VQH6 E5L9R9 A0A3M6T989 A0A2P2I560 G1KPH1
PDB
4KLZ
E-value=1.19069e-56,
Score=552
Ontologies
GO
GO:0007165
GO:0003924
GO:0016020
GO:0005525
GO:0016021
GO:0009408
GO:0006970
GO:0006979
GO:0042594
GO:0005516
GO:0019239
GO:0007265
GO:0005886
GO:0019003
GO:0004767
GO:0045944
GO:0030215
GO:0003682
GO:0044297
GO:0010977
GO:0010976
GO:0097447
GO:0032507
GO:0045121
GO:0032489
GO:0005634
GO:0005737
GO:0030100
GO:0007264
GO:0006468
GO:0006811
GO:0043564
GO:0006418
GO:0003824
PANTHER
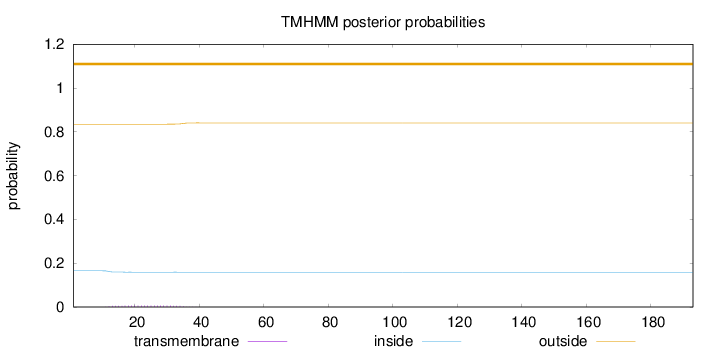
Topology
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13742
Exp number, first 60 AAs:
0.13684
Total prob of N-in:
0.16506
outside
1 - 193
Population Genetic Test Statistics
Pi
231.28582
Theta
227.290535
Tajima's D
1.0477
CLR
0
CSRT
0.673116344182791
Interpretation
Uncertain
