Gene
KWMTBOMO11673
Pre Gene Modal
BGIBMGA003981
Annotation
PREDICTED:_atrial_natriuretic_peptide_receptor_1_[Bombyx_mori]
Full name
Guanylate cyclase
Location in the cell
PlasmaMembrane Reliability : 1.781
Sequence
CDS
ATGATCATCTTTACGTGTGTATTGGTCTTGCCGATGTTGTCCGGTACGGTTTTAAACAAAACTGTATACAAAATAGGGGTCTTAATGGCCTCTCGGTTGGACTCGCCTTTTGACTTGGAAAGATGTGGACCGGCTGTGGACCTGGGACTTAAGAAAGTGAATGAAAAGTTTCTCTCGCATCACGGAATTGAGTTGGAGAAGGTTCAAGGAAGTTACCCATCGTGCTCGGGTGCCATAGCTCCGGGTTTGGCCGCCGACATGCACTTCAAGAACGACGTGATCGCTTTCATAGGACCCGCGTGCGCGTTCGCCTTGGAACCGGTTGCGAGGCTGGCCGCTTACTGGAATACTCCCATCATCACCGGAATGGGGGATCAGATAGTTCAGATTTAA
Protein
MIIFTCVLVLPMLSGTVLNKTVYKIGVLMASRLDSPFDLERCGPAVDLGLKKVNEKFLSHHGIELEKVQGSYPSCSGAIAPGLAADMHFKNDVIAFIGPACAFALEPVARLAAYWNTPIITGMGDQIVQI
Summary
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Feature
chain Guanylate cyclase
Uniprot
A0A2A4JER9
A0A0L7LM81
H9J394
A0A2H1WLK9
A0A0N1IPJ9
A0A194QCN7
+ More
A0A232EQ27 A0A212FAS7 D6WRM6 A0A194QY55 A0A1I8QAW1 A0A034VK64 A0A0M4E8Y6 A0A0Q9XIU0 A0A0Q9WDQ8 A0A182MJD1 A0A1I8MX63 A0A336KPL4 B4J8X4 A0A1B0G3T4 B4LQ69 B4KRH8 A0A3L8DR11 A0A1A9ZVI9 A0A1B0BZE8 A0A0L0C030 A0A182JJG6 A0A2S2PGL0 A0A151J3M2 A0A158NDH8 A0A195B5P1 A0A195F339 A0A0R1DS54 B4P9K3 A0A1W4V6D9 A0A154P996 A0A0P9BWM0 A0A2J7QWW5 B3MIB2 K7JAC0 B4NN48 A0A0Q5W083 A0A3B0JUU0 Q28X38 B4H4Z8 B3NK16 A0A310S5S1 A0A067RK75 A0A182KYH8 A0A0N0BDY8 Q9W2P1 A0A0J9RIQ1 B0W1T4 A0A2A3E503 A0A182QDM9 A0A2S2Q6P6 B4I7H3 Q8MLX0 B4QFJ8 A0A0J9U744 A0A2H8TER7 J9K9Z0 A0A182TFP5 A0A084VCP6 A0A087ZY46 A0A026WAY9 F4W994 A0A182GFN3 U4UHB1 A0A182P368 A0A151X8Y4 A0A182VTD2 E2APH5 A0A0L7RCA0 E9J755 A0A1S4GYZ1 A0A182F3A0 A0A151IAC0 W5JE51 Q7Q8C9 A0A226F360 A0A182VNR5 A0A1D2MPH3 R7TF11 V4ALG5 R7T4L4 A0A2P6KSU1 A0A087UZV2 A0A2T7PJ64 R7TBU9 T1KEH1
A0A232EQ27 A0A212FAS7 D6WRM6 A0A194QY55 A0A1I8QAW1 A0A034VK64 A0A0M4E8Y6 A0A0Q9XIU0 A0A0Q9WDQ8 A0A182MJD1 A0A1I8MX63 A0A336KPL4 B4J8X4 A0A1B0G3T4 B4LQ69 B4KRH8 A0A3L8DR11 A0A1A9ZVI9 A0A1B0BZE8 A0A0L0C030 A0A182JJG6 A0A2S2PGL0 A0A151J3M2 A0A158NDH8 A0A195B5P1 A0A195F339 A0A0R1DS54 B4P9K3 A0A1W4V6D9 A0A154P996 A0A0P9BWM0 A0A2J7QWW5 B3MIB2 K7JAC0 B4NN48 A0A0Q5W083 A0A3B0JUU0 Q28X38 B4H4Z8 B3NK16 A0A310S5S1 A0A067RK75 A0A182KYH8 A0A0N0BDY8 Q9W2P1 A0A0J9RIQ1 B0W1T4 A0A2A3E503 A0A182QDM9 A0A2S2Q6P6 B4I7H3 Q8MLX0 B4QFJ8 A0A0J9U744 A0A2H8TER7 J9K9Z0 A0A182TFP5 A0A084VCP6 A0A087ZY46 A0A026WAY9 F4W994 A0A182GFN3 U4UHB1 A0A182P368 A0A151X8Y4 A0A182VTD2 E2APH5 A0A0L7RCA0 E9J755 A0A1S4GYZ1 A0A182F3A0 A0A151IAC0 W5JE51 Q7Q8C9 A0A226F360 A0A182VNR5 A0A1D2MPH3 R7TF11 V4ALG5 R7T4L4 A0A2P6KSU1 A0A087UZV2 A0A2T7PJ64 R7TBU9 T1KEH1
EC Number
4.6.1.2
Pubmed
26227816
19121390
26354079
28648823
22118469
18362917
+ More
19820115 25348373 17994087 25315136 30249741 26108605 21347285 17550304 20075255 15632085 24845553 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 24438588 24508170 21719571 26483478 23537049 20798317 21282665 12364791 20920257 23761445 27289101 23254933
19820115 25348373 17994087 25315136 30249741 26108605 21347285 17550304 20075255 15632085 24845553 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 24438588 24508170 21719571 26483478 23537049 20798317 21282665 12364791 20920257 23761445 27289101 23254933
EMBL
NWSH01001857
PCG69902.1
JTDY01000654
KOB76321.1
BABH01021725
ODYU01009488
+ More
SOQ53961.1 KQ460459 KPJ14653.1 KQ459185 KPJ03232.1 NNAY01002859 OXU20432.1 AGBW02009432 OWR50831.1 KQ971354 EFA07684.2 KQ460949 KPJ10473.1 GAKP01016440 GAKP01016438 JAC42512.1 CP012524 ALC41271.1 CH933808 KRG05220.1 CH940648 KRF79937.1 AXCM01008450 UFQS01000670 UFQT01000670 SSX05992.1 SSX26349.1 CH916367 EDW01323.1 CCAG010019265 EDW61354.2 EDW10404.2 QOIP01000005 RLU22831.1 JXJN01023105 JRES01001096 KNC25591.1 GGMR01015962 MBY28581.1 KQ980275 KYN17032.1 ADTU01001036 ADTU01001037 KQ976598 KYM79590.1 KQ981855 KYN34878.1 CM000158 KRJ99806.1 EDW91323.1 KQ434829 KZC07740.1 CH902619 KPU75902.1 NEVH01009413 PNF33087.1 EDV35957.1 AAZX01000434 AAZX01000435 AAZX01012324 AAZX01016843 CH964282 EDW85787.1 CH954179 KQS62239.1 OUUW01000001 SPP74838.1 CM000071 EAL26478.3 CH479210 EDW32834.1 EDV55382.1 KQ769179 OAD52914.1 KK852461 KDR23428.1 KQ435847 KOX71134.1 AE013599 AAF46649.2 CM002911 KMY95374.1 DS231823 EDS26635.1 KZ288369 PBC26770.1 AXCN02002197 GGMS01003659 MBY72862.1 CH480824 EDW56548.1 AAM68187.2 CM000362 EDX07990.1 KMY95375.1 GFXV01000769 MBW12574.1 ABLF02027323 ABLF02027327 ABLF02027333 ABLF02027334 ABLF02027335 ATLV01010838 KE524620 KFB35740.1 KK107293 EZA53272.1 GL888002 EGI69276.1 JXUM01060110 JXUM01060111 JXUM01060112 JXUM01060113 JXUM01060114 KQ562088 KXJ76720.1 KB632300 ERL91753.1 KQ982402 KYQ56837.1 GL441542 EFN64674.1 KQ414616 KOC68458.1 GL768421 EFZ11362.1 AAAB01008944 KQ978223 KYM96262.1 ADMH02001784 ETN61094.1 EAA10186.4 LNIX01000001 OXA63366.1 LJIJ01000725 ODM94989.1 AMQN01002821 AMQN01002822 KB310159 ELT92328.1 KB201304 ESO97952.1 AMQN01003544 KB312101 ELT87878.1 MWRG01005805 PRD29395.1 KK122519 KFM82891.1 PZQS01000003 PVD33471.1 AMQN01002981 AMQN01002982 KB310616 ELT91198.1 CAEY01002035
SOQ53961.1 KQ460459 KPJ14653.1 KQ459185 KPJ03232.1 NNAY01002859 OXU20432.1 AGBW02009432 OWR50831.1 KQ971354 EFA07684.2 KQ460949 KPJ10473.1 GAKP01016440 GAKP01016438 JAC42512.1 CP012524 ALC41271.1 CH933808 KRG05220.1 CH940648 KRF79937.1 AXCM01008450 UFQS01000670 UFQT01000670 SSX05992.1 SSX26349.1 CH916367 EDW01323.1 CCAG010019265 EDW61354.2 EDW10404.2 QOIP01000005 RLU22831.1 JXJN01023105 JRES01001096 KNC25591.1 GGMR01015962 MBY28581.1 KQ980275 KYN17032.1 ADTU01001036 ADTU01001037 KQ976598 KYM79590.1 KQ981855 KYN34878.1 CM000158 KRJ99806.1 EDW91323.1 KQ434829 KZC07740.1 CH902619 KPU75902.1 NEVH01009413 PNF33087.1 EDV35957.1 AAZX01000434 AAZX01000435 AAZX01012324 AAZX01016843 CH964282 EDW85787.1 CH954179 KQS62239.1 OUUW01000001 SPP74838.1 CM000071 EAL26478.3 CH479210 EDW32834.1 EDV55382.1 KQ769179 OAD52914.1 KK852461 KDR23428.1 KQ435847 KOX71134.1 AE013599 AAF46649.2 CM002911 KMY95374.1 DS231823 EDS26635.1 KZ288369 PBC26770.1 AXCN02002197 GGMS01003659 MBY72862.1 CH480824 EDW56548.1 AAM68187.2 CM000362 EDX07990.1 KMY95375.1 GFXV01000769 MBW12574.1 ABLF02027323 ABLF02027327 ABLF02027333 ABLF02027334 ABLF02027335 ATLV01010838 KE524620 KFB35740.1 KK107293 EZA53272.1 GL888002 EGI69276.1 JXUM01060110 JXUM01060111 JXUM01060112 JXUM01060113 JXUM01060114 KQ562088 KXJ76720.1 KB632300 ERL91753.1 KQ982402 KYQ56837.1 GL441542 EFN64674.1 KQ414616 KOC68458.1 GL768421 EFZ11362.1 AAAB01008944 KQ978223 KYM96262.1 ADMH02001784 ETN61094.1 EAA10186.4 LNIX01000001 OXA63366.1 LJIJ01000725 ODM94989.1 AMQN01002821 AMQN01002822 KB310159 ELT92328.1 KB201304 ESO97952.1 AMQN01003544 KB312101 ELT87878.1 MWRG01005805 PRD29395.1 KK122519 KFM82891.1 PZQS01000003 PVD33471.1 AMQN01002981 AMQN01002982 KB310616 ELT91198.1 CAEY01002035
Proteomes
UP000218220
UP000037510
UP000005204
UP000053240
UP000053268
UP000215335
+ More
UP000007151 UP000007266 UP000095300 UP000092553 UP000009192 UP000008792 UP000075883 UP000095301 UP000001070 UP000092444 UP000279307 UP000092445 UP000092460 UP000037069 UP000075880 UP000078492 UP000005205 UP000078540 UP000078541 UP000002282 UP000192221 UP000076502 UP000007801 UP000235965 UP000002358 UP000007798 UP000008711 UP000268350 UP000001819 UP000008744 UP000027135 UP000075882 UP000053105 UP000000803 UP000002320 UP000242457 UP000075886 UP000001292 UP000000304 UP000007819 UP000075902 UP000030765 UP000005203 UP000053097 UP000007755 UP000069940 UP000249989 UP000030742 UP000075885 UP000075809 UP000075920 UP000000311 UP000053825 UP000069272 UP000078542 UP000000673 UP000007062 UP000198287 UP000075903 UP000094527 UP000014760 UP000030746 UP000054359 UP000245119 UP000015104
UP000007151 UP000007266 UP000095300 UP000092553 UP000009192 UP000008792 UP000075883 UP000095301 UP000001070 UP000092444 UP000279307 UP000092445 UP000092460 UP000037069 UP000075880 UP000078492 UP000005205 UP000078540 UP000078541 UP000002282 UP000192221 UP000076502 UP000007801 UP000235965 UP000002358 UP000007798 UP000008711 UP000268350 UP000001819 UP000008744 UP000027135 UP000075882 UP000053105 UP000000803 UP000002320 UP000242457 UP000075886 UP000001292 UP000000304 UP000007819 UP000075902 UP000030765 UP000005203 UP000053097 UP000007755 UP000069940 UP000249989 UP000030742 UP000075885 UP000075809 UP000075920 UP000000311 UP000053825 UP000069272 UP000078542 UP000000673 UP000007062 UP000198287 UP000075903 UP000094527 UP000014760 UP000030746 UP000054359 UP000245119 UP000015104
Pfam
Interpro
IPR028082
Peripla_BP_I
+ More
IPR001828 ANF_lig-bd_rcpt
IPR001170 ANPR/GUC
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR001054 A/G_cyclase
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR018297 A/G_cyclase_CS
IPR011645 HNOB_dom_associated
IPR004843 Calcineurin-like_PHP_ApaH
IPR041871 MPP_TMEM62
IPR029052 Metallo-depent_PP-like
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR001828 ANF_lig-bd_rcpt
IPR001170 ANPR/GUC
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR001054 A/G_cyclase
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR018297 A/G_cyclase_CS
IPR011645 HNOB_dom_associated
IPR004843 Calcineurin-like_PHP_ApaH
IPR041871 MPP_TMEM62
IPR029052 Metallo-depent_PP-like
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
Gene 3D
ProteinModelPortal
A0A2A4JER9
A0A0L7LM81
H9J394
A0A2H1WLK9
A0A0N1IPJ9
A0A194QCN7
+ More
A0A232EQ27 A0A212FAS7 D6WRM6 A0A194QY55 A0A1I8QAW1 A0A034VK64 A0A0M4E8Y6 A0A0Q9XIU0 A0A0Q9WDQ8 A0A182MJD1 A0A1I8MX63 A0A336KPL4 B4J8X4 A0A1B0G3T4 B4LQ69 B4KRH8 A0A3L8DR11 A0A1A9ZVI9 A0A1B0BZE8 A0A0L0C030 A0A182JJG6 A0A2S2PGL0 A0A151J3M2 A0A158NDH8 A0A195B5P1 A0A195F339 A0A0R1DS54 B4P9K3 A0A1W4V6D9 A0A154P996 A0A0P9BWM0 A0A2J7QWW5 B3MIB2 K7JAC0 B4NN48 A0A0Q5W083 A0A3B0JUU0 Q28X38 B4H4Z8 B3NK16 A0A310S5S1 A0A067RK75 A0A182KYH8 A0A0N0BDY8 Q9W2P1 A0A0J9RIQ1 B0W1T4 A0A2A3E503 A0A182QDM9 A0A2S2Q6P6 B4I7H3 Q8MLX0 B4QFJ8 A0A0J9U744 A0A2H8TER7 J9K9Z0 A0A182TFP5 A0A084VCP6 A0A087ZY46 A0A026WAY9 F4W994 A0A182GFN3 U4UHB1 A0A182P368 A0A151X8Y4 A0A182VTD2 E2APH5 A0A0L7RCA0 E9J755 A0A1S4GYZ1 A0A182F3A0 A0A151IAC0 W5JE51 Q7Q8C9 A0A226F360 A0A182VNR5 A0A1D2MPH3 R7TF11 V4ALG5 R7T4L4 A0A2P6KSU1 A0A087UZV2 A0A2T7PJ64 R7TBU9 T1KEH1
A0A232EQ27 A0A212FAS7 D6WRM6 A0A194QY55 A0A1I8QAW1 A0A034VK64 A0A0M4E8Y6 A0A0Q9XIU0 A0A0Q9WDQ8 A0A182MJD1 A0A1I8MX63 A0A336KPL4 B4J8X4 A0A1B0G3T4 B4LQ69 B4KRH8 A0A3L8DR11 A0A1A9ZVI9 A0A1B0BZE8 A0A0L0C030 A0A182JJG6 A0A2S2PGL0 A0A151J3M2 A0A158NDH8 A0A195B5P1 A0A195F339 A0A0R1DS54 B4P9K3 A0A1W4V6D9 A0A154P996 A0A0P9BWM0 A0A2J7QWW5 B3MIB2 K7JAC0 B4NN48 A0A0Q5W083 A0A3B0JUU0 Q28X38 B4H4Z8 B3NK16 A0A310S5S1 A0A067RK75 A0A182KYH8 A0A0N0BDY8 Q9W2P1 A0A0J9RIQ1 B0W1T4 A0A2A3E503 A0A182QDM9 A0A2S2Q6P6 B4I7H3 Q8MLX0 B4QFJ8 A0A0J9U744 A0A2H8TER7 J9K9Z0 A0A182TFP5 A0A084VCP6 A0A087ZY46 A0A026WAY9 F4W994 A0A182GFN3 U4UHB1 A0A182P368 A0A151X8Y4 A0A182VTD2 E2APH5 A0A0L7RCA0 E9J755 A0A1S4GYZ1 A0A182F3A0 A0A151IAC0 W5JE51 Q7Q8C9 A0A226F360 A0A182VNR5 A0A1D2MPH3 R7TF11 V4ALG5 R7T4L4 A0A2P6KSU1 A0A087UZV2 A0A2T7PJ64 R7TBU9 T1KEH1
PDB
3A3K
E-value=8.7253e-10,
Score=145
Ontologies
PATHWAY
GO
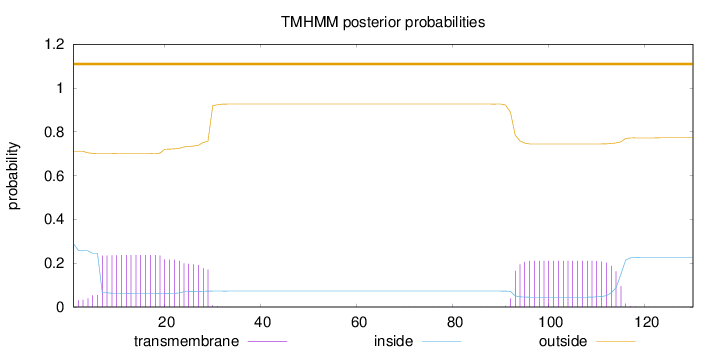
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.691846
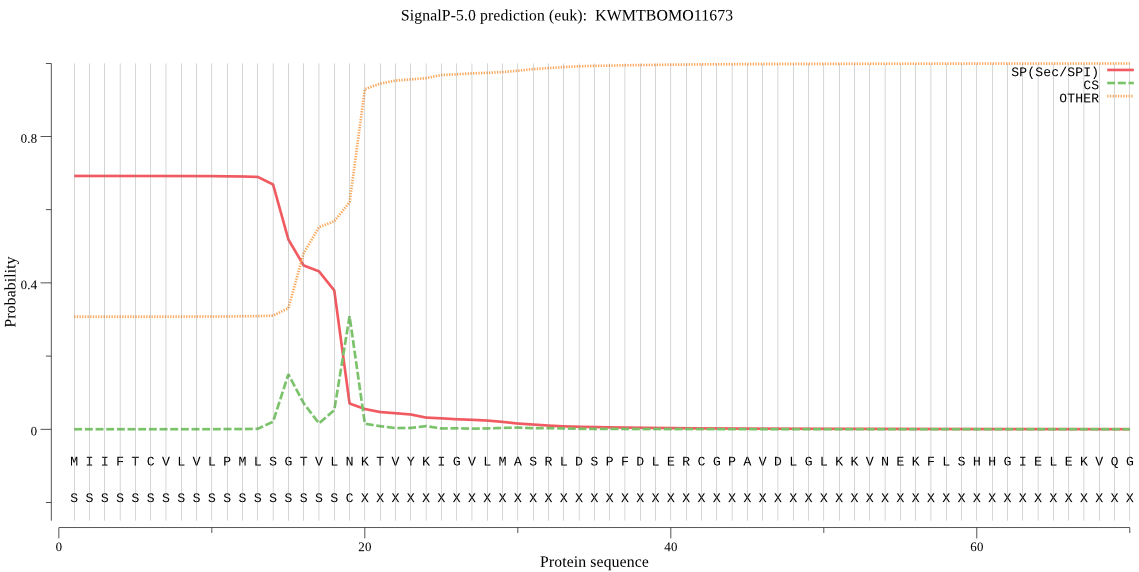
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.94506
Exp number, first 60 AAs:
5.28285
Total prob of N-in:
0.28882
outside
1 - 130
Population Genetic Test Statistics
Pi
337.044149
Theta
201.747059
Tajima's D
2.110982
CLR
0.383907
CSRT
0.904604769761512
Interpretation
Uncertain
