Gene
KWMTBOMO11659
Pre Gene Modal
BGIBMGA004130
Annotation
Malate_dehydrogenase_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.294
Sequence
CDS
ATGCCTGAGGTACTAGTTGAGGAAGCCAGGCGGTTCATGGAGGACAGCCTCACGGCTGTAGGCGCTCCTGTATCCGAAGCGAAAGCGCAAGCTGATCTTCTAATCCATGCTGATACAGTCGGACATTACAGTCACGGACTTAACAGATTGGAATTCTATATAAACGATATCCTGTCAAAAGCAACAGATCCTTGTGCAAAGCCGGTGATATTGAAAGAGTCGGCTGCGACCGCTCTGGTGGACGGTTGCGACGCTTTAGGTGCTACTGTCGGAAACTTCTGTATGGAAGTCGCGATCAGGAAGGCGAAGGAAGCGGGAGTAGGATGGGTGGCTGCCAGACGTAGCAATCATTACGGTATGGCTGGATACTGGGCATTGAAGGCGGAAAAACAGGGTCTCATAGGACTGTCGTTTACAAATTCTTCCCCGATATTAGTACCGACTAGATCTAAAACAAGTGCATTGGGTACAAATCCCATAGCTTTGGCCGCTCCAGCTAAAGACGGCGATAATTTGGTTGTAGATTTAGCTACAACGGCTGTTGCTATGGGCAAAGTTGAAATACAAGTACACAAAGAGGAGCCATTGCCAGCTGGCTGGGCGCTCGGCCCCGATGGAAAAACTACCACTGATGCTAAATTGGCTTTCGAAACAGCTAAACTCCTGCCGCTGGGAGGTGCTGAAGAGACCAGTGGTTACAAAGGGTATGCTTTGAGCGCCCTTGTTGAAGTATTGTGCAGTGGATTATCGGGATCTAAATCGACGCATCAAGTCCGTGGTTGGGAACTGGACAGTCAAGGCGGACCTCCGGATCTGGGGCACTGCTTCGCGGCCATCGACCCGGGCTGCTTCGCTCCTGGCTTCGAAGACAGGATCTCGAACTGCTTACAGCACTGGAGGAACTTAGAACCGATGGACCCTAAACTCCCCGTACTAGCACCAGGAGACAAAGAACGCAAGAACTACGCGGAAACACAGAAGAGAGGAACCATAAACTACCCTCAGAAACAGTACGAAGCTTACTTCAAAATGGCGAATCGAATCGGAATAAAACCAATGCAGATAGTGTGA
Protein
MPEVLVEEARRFMEDSLTAVGAPVSEAKAQADLLIHADTVGHYSHGLNRLEFYINDILSKATDPCAKPVILKESAATALVDGCDALGATVGNFCMEVAIRKAKEAGVGWVAARRSNHYGMAGYWALKAEKQGLIGLSFTNSSPILVPTRSKTSALGTNPIALAAPAKDGDNLVVDLATTAVAMGKVEIQVHKEEPLPAGWALGPDGKTTTDAKLAFETAKLLPLGGAEETSGYKGYALSALVEVLCSGLSGSKSTHQVRGWELDSQGGPPDLGHCFAAIDPGCFAPGFEDRISNCLQHWRNLEPMDPKLPVLAPGDKERKNYAETQKRGTINYPQKQYEAYFKMANRIGIKPMQIV
Summary
Uniprot
H9J3P3
A0A2H1VF87
A0A2H1VG35
A0A2A4JT31
A0A194QIG5
S4PYB2
+ More
A0A212FNJ9 A0A3S2NAY3 A0A194QCP7 A0A212FNQ3 A0A2H1VFV1 A0A1B6MIR7 A0A194QYE0 A0A084WMQ8 T1E821 A0A2A4JTQ6 W5JGY0 A0A182J993 A0A182TC41 A0A2M4A9B8 A0A2M4A8Y9 A0A3F2Z255 A0A2M4A993 Q7PI68 A0A2M4BMC1 A0A2M3Z812 A0A3F2YSU0 A0A2M4BLN2 A0A182UET8 A0A182KC31 A0A182MF34 A0A3F2YUT7 A0A182PDI9 A0A3F2Z243 A0A182V6K4 A7UUE0 A0A182QHD3 A0A3F2YSU4 A0A182NIS9 A0A1Q3FLN1 A0A182Y2M5 A0A2J7REG8 B0XBA9 A0A2J7REG9 B4LHA3 A0A1Y1NQ32 B4K0I1 A0A0Q9WLQ0 A0A0Q9WS29 B4J137 A0A182GX03 A0A2A4JUX8 A0A194R3R2 A0A336KKY3 U5ET27 B4H5Y0 B4ML58 A0A336M4M8 A0A3B0KNF1 A0A2A4JUW3 Q8IPT9 A0A0Q9XME1 A0A3B0JTA1 A0A0M3QX38 B4KUL3 Q2LYQ7 Q8IPU0 B3NIW8 A0A1W4V9A7 A0A0J9RZA6 A0A182FEX8 Q9VP68 A0A182RDI2 A0A0J9S0M4 A0A1L8E132 B4ITH5 A0A0P8Y9U6 B4PE63 B5RIN9 A0A0K8TRR8 B4IAK9 B3M6T1 A0A1D2NDY4 Q961U7 A0A034VHT6 A0A182WBT0 A0A1L8E1J8 A0A034VM42 W8B513 A0A0K8UVR2 W8B112 A0A1J1IRZ0 A0A0A1XSQ8 A0A0A1WKR5 A0A0A1WH71 A0A3R7PNK7 A0A1B0ABA4 A0A1B6H9E9 W5LY39 W5LY58
A0A212FNJ9 A0A3S2NAY3 A0A194QCP7 A0A212FNQ3 A0A2H1VFV1 A0A1B6MIR7 A0A194QYE0 A0A084WMQ8 T1E821 A0A2A4JTQ6 W5JGY0 A0A182J993 A0A182TC41 A0A2M4A9B8 A0A2M4A8Y9 A0A3F2Z255 A0A2M4A993 Q7PI68 A0A2M4BMC1 A0A2M3Z812 A0A3F2YSU0 A0A2M4BLN2 A0A182UET8 A0A182KC31 A0A182MF34 A0A3F2YUT7 A0A182PDI9 A0A3F2Z243 A0A182V6K4 A7UUE0 A0A182QHD3 A0A3F2YSU4 A0A182NIS9 A0A1Q3FLN1 A0A182Y2M5 A0A2J7REG8 B0XBA9 A0A2J7REG9 B4LHA3 A0A1Y1NQ32 B4K0I1 A0A0Q9WLQ0 A0A0Q9WS29 B4J137 A0A182GX03 A0A2A4JUX8 A0A194R3R2 A0A336KKY3 U5ET27 B4H5Y0 B4ML58 A0A336M4M8 A0A3B0KNF1 A0A2A4JUW3 Q8IPT9 A0A0Q9XME1 A0A3B0JTA1 A0A0M3QX38 B4KUL3 Q2LYQ7 Q8IPU0 B3NIW8 A0A1W4V9A7 A0A0J9RZA6 A0A182FEX8 Q9VP68 A0A182RDI2 A0A0J9S0M4 A0A1L8E132 B4ITH5 A0A0P8Y9U6 B4PE63 B5RIN9 A0A0K8TRR8 B4IAK9 B3M6T1 A0A1D2NDY4 Q961U7 A0A034VHT6 A0A182WBT0 A0A1L8E1J8 A0A034VM42 W8B513 A0A0K8UVR2 W8B112 A0A1J1IRZ0 A0A0A1XSQ8 A0A0A1WKR5 A0A0A1WH71 A0A3R7PNK7 A0A1B0ABA4 A0A1B6H9E9 W5LY39 W5LY58
Pubmed
19121390
26354079
23622113
22118469
24438588
20920257
+ More
23761445 12364791 14747013 17210077 20966253 25244985 17994087 28004739 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249 17550304 26369729 27289101 25348373 24495485 25830018
23761445 12364791 14747013 17210077 20966253 25244985 17994087 28004739 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249 17550304 26369729 27289101 25348373 24495485 25830018
EMBL
BABH01021703
ODYU01002232
SOQ39441.1
ODYU01002355
SOQ39741.1
NWSH01000623
+ More
PCG75197.1 KQ459185 KPJ03241.1 GAIX01003593 JAA88967.1 AGBW02005277 OWR55321.1 RSAL01000136 RVE46247.1 KPJ03242.1 OWR55320.1 SOQ39740.1 GEBQ01004139 JAT35838.1 KQ460949 KPJ10482.1 ATLV01024501 KE525352 KFB51502.1 GAMD01003123 JAA98467.1 PCG75199.1 ADMH02001558 ETN62069.1 GGFK01004075 MBW37396.1 GGFK01003936 MBW37257.1 GGFK01004053 MBW37374.1 AAAB01008960 EAA44260.4 GGFJ01004950 MBW54091.1 GGFM01003906 MBW24657.1 APCN01002308 GGFJ01004801 MBW53942.1 AXCM01005053 EDO63918.1 EDO63919.1 AXCN02000480 GFDL01006600 JAV28445.1 NEVH01004981 PNF39233.1 DS232623 EDS44201.1 PNF39234.1 CH940647 EDW70616.1 GEZM01000243 JAV98447.1 CH916456 EDV98661.1 KRF85014.1 KRF85015.1 CH916366 EDV96892.1 JXUM01020208 JXUM01020209 KQ560566 KXJ81871.1 PCG75202.1 KPJ10481.1 UFQS01000523 UFQT01000523 SSX04578.1 SSX24941.1 GANO01002123 JAB57748.1 CH479212 EDW33197.1 CH963847 EDW73116.2 SSX04577.1 SSX24940.1 OUUW01000009 SPP85348.1 PCG75200.1 AE014296 BT150433 AAN12159.1 AHL21637.1 CH933809 KRG06078.1 SPP85347.1 CP012525 ALC44997.1 ALC45189.1 EDW18241.1 CH379069 EAL29804.2 AAN12158.1 CH954178 EDV52614.1 CM002912 KMZ00944.1 AAF51687.1 KMZ00945.1 GFDF01001710 JAV12374.1 CH891699 EDW99688.1 CH902618 KPU78208.1 CM000159 EDW95076.1 BT044163 ACH92228.1 GDAI01001013 JAI16590.1 CH480826 EDW44322.1 EDV39767.1 LJIJ01000072 ODN03478.1 AY047551 AAK77283.1 GAKP01016086 JAC42866.1 GFDF01001709 JAV12375.1 GAKP01016087 JAC42865.1 GAMC01014377 GAMC01014375 JAB92178.1 GDHF01031803 GDHF01021560 JAI20511.1 JAI30754.1 GAMC01014378 GAMC01014376 JAB92177.1 CVRI01000058 CRL02920.1 GBXI01000704 JAD13588.1 GBXI01014658 JAC99633.1 GBXI01016549 JAC97742.1 QCYY01001490 ROT77718.1 GECU01036368 JAS71338.1 AHAT01008084 AHAT01008085
PCG75197.1 KQ459185 KPJ03241.1 GAIX01003593 JAA88967.1 AGBW02005277 OWR55321.1 RSAL01000136 RVE46247.1 KPJ03242.1 OWR55320.1 SOQ39740.1 GEBQ01004139 JAT35838.1 KQ460949 KPJ10482.1 ATLV01024501 KE525352 KFB51502.1 GAMD01003123 JAA98467.1 PCG75199.1 ADMH02001558 ETN62069.1 GGFK01004075 MBW37396.1 GGFK01003936 MBW37257.1 GGFK01004053 MBW37374.1 AAAB01008960 EAA44260.4 GGFJ01004950 MBW54091.1 GGFM01003906 MBW24657.1 APCN01002308 GGFJ01004801 MBW53942.1 AXCM01005053 EDO63918.1 EDO63919.1 AXCN02000480 GFDL01006600 JAV28445.1 NEVH01004981 PNF39233.1 DS232623 EDS44201.1 PNF39234.1 CH940647 EDW70616.1 GEZM01000243 JAV98447.1 CH916456 EDV98661.1 KRF85014.1 KRF85015.1 CH916366 EDV96892.1 JXUM01020208 JXUM01020209 KQ560566 KXJ81871.1 PCG75202.1 KPJ10481.1 UFQS01000523 UFQT01000523 SSX04578.1 SSX24941.1 GANO01002123 JAB57748.1 CH479212 EDW33197.1 CH963847 EDW73116.2 SSX04577.1 SSX24940.1 OUUW01000009 SPP85348.1 PCG75200.1 AE014296 BT150433 AAN12159.1 AHL21637.1 CH933809 KRG06078.1 SPP85347.1 CP012525 ALC44997.1 ALC45189.1 EDW18241.1 CH379069 EAL29804.2 AAN12158.1 CH954178 EDV52614.1 CM002912 KMZ00944.1 AAF51687.1 KMZ00945.1 GFDF01001710 JAV12374.1 CH891699 EDW99688.1 CH902618 KPU78208.1 CM000159 EDW95076.1 BT044163 ACH92228.1 GDAI01001013 JAI16590.1 CH480826 EDW44322.1 EDV39767.1 LJIJ01000072 ODN03478.1 AY047551 AAK77283.1 GAKP01016086 JAC42866.1 GFDF01001709 JAV12375.1 GAKP01016087 JAC42865.1 GAMC01014377 GAMC01014375 JAB92178.1 GDHF01031803 GDHF01021560 JAI20511.1 JAI30754.1 GAMC01014378 GAMC01014376 JAB92177.1 CVRI01000058 CRL02920.1 GBXI01000704 JAD13588.1 GBXI01014658 JAC99633.1 GBXI01016549 JAC97742.1 QCYY01001490 ROT77718.1 GECU01036368 JAS71338.1 AHAT01008084 AHAT01008085
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000283053
UP000053240
+ More
UP000030765 UP000000673 UP000075880 UP000075901 UP000076407 UP000007062 UP000075840 UP000075902 UP000075881 UP000075883 UP000075882 UP000075885 UP000075903 UP000075886 UP000075884 UP000076408 UP000235965 UP000002320 UP000008792 UP000001070 UP000069940 UP000249989 UP000008744 UP000007798 UP000268350 UP000000803 UP000009192 UP000092553 UP000001819 UP000008711 UP000192221 UP000069272 UP000075900 UP000002282 UP000007801 UP000001292 UP000094527 UP000075920 UP000183832 UP000283509 UP000092445 UP000018468
UP000030765 UP000000673 UP000075880 UP000075901 UP000076407 UP000007062 UP000075840 UP000075902 UP000075881 UP000075883 UP000075882 UP000075885 UP000075903 UP000075886 UP000075884 UP000076408 UP000235965 UP000002320 UP000008792 UP000001070 UP000069940 UP000249989 UP000008744 UP000007798 UP000268350 UP000000803 UP000009192 UP000092553 UP000001819 UP000008711 UP000192221 UP000069272 UP000075900 UP000002282 UP000007801 UP000001292 UP000094527 UP000075920 UP000183832 UP000283509 UP000092445 UP000018468
Pfam
Interpro
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J3P3
A0A2H1VF87
A0A2H1VG35
A0A2A4JT31
A0A194QIG5
S4PYB2
+ More
A0A212FNJ9 A0A3S2NAY3 A0A194QCP7 A0A212FNQ3 A0A2H1VFV1 A0A1B6MIR7 A0A194QYE0 A0A084WMQ8 T1E821 A0A2A4JTQ6 W5JGY0 A0A182J993 A0A182TC41 A0A2M4A9B8 A0A2M4A8Y9 A0A3F2Z255 A0A2M4A993 Q7PI68 A0A2M4BMC1 A0A2M3Z812 A0A3F2YSU0 A0A2M4BLN2 A0A182UET8 A0A182KC31 A0A182MF34 A0A3F2YUT7 A0A182PDI9 A0A3F2Z243 A0A182V6K4 A7UUE0 A0A182QHD3 A0A3F2YSU4 A0A182NIS9 A0A1Q3FLN1 A0A182Y2M5 A0A2J7REG8 B0XBA9 A0A2J7REG9 B4LHA3 A0A1Y1NQ32 B4K0I1 A0A0Q9WLQ0 A0A0Q9WS29 B4J137 A0A182GX03 A0A2A4JUX8 A0A194R3R2 A0A336KKY3 U5ET27 B4H5Y0 B4ML58 A0A336M4M8 A0A3B0KNF1 A0A2A4JUW3 Q8IPT9 A0A0Q9XME1 A0A3B0JTA1 A0A0M3QX38 B4KUL3 Q2LYQ7 Q8IPU0 B3NIW8 A0A1W4V9A7 A0A0J9RZA6 A0A182FEX8 Q9VP68 A0A182RDI2 A0A0J9S0M4 A0A1L8E132 B4ITH5 A0A0P8Y9U6 B4PE63 B5RIN9 A0A0K8TRR8 B4IAK9 B3M6T1 A0A1D2NDY4 Q961U7 A0A034VHT6 A0A182WBT0 A0A1L8E1J8 A0A034VM42 W8B513 A0A0K8UVR2 W8B112 A0A1J1IRZ0 A0A0A1XSQ8 A0A0A1WKR5 A0A0A1WH71 A0A3R7PNK7 A0A1B0ABA4 A0A1B6H9E9 W5LY39 W5LY58
A0A212FNJ9 A0A3S2NAY3 A0A194QCP7 A0A212FNQ3 A0A2H1VFV1 A0A1B6MIR7 A0A194QYE0 A0A084WMQ8 T1E821 A0A2A4JTQ6 W5JGY0 A0A182J993 A0A182TC41 A0A2M4A9B8 A0A2M4A8Y9 A0A3F2Z255 A0A2M4A993 Q7PI68 A0A2M4BMC1 A0A2M3Z812 A0A3F2YSU0 A0A2M4BLN2 A0A182UET8 A0A182KC31 A0A182MF34 A0A3F2YUT7 A0A182PDI9 A0A3F2Z243 A0A182V6K4 A7UUE0 A0A182QHD3 A0A3F2YSU4 A0A182NIS9 A0A1Q3FLN1 A0A182Y2M5 A0A2J7REG8 B0XBA9 A0A2J7REG9 B4LHA3 A0A1Y1NQ32 B4K0I1 A0A0Q9WLQ0 A0A0Q9WS29 B4J137 A0A182GX03 A0A2A4JUX8 A0A194R3R2 A0A336KKY3 U5ET27 B4H5Y0 B4ML58 A0A336M4M8 A0A3B0KNF1 A0A2A4JUW3 Q8IPT9 A0A0Q9XME1 A0A3B0JTA1 A0A0M3QX38 B4KUL3 Q2LYQ7 Q8IPU0 B3NIW8 A0A1W4V9A7 A0A0J9RZA6 A0A182FEX8 Q9VP68 A0A182RDI2 A0A0J9S0M4 A0A1L8E132 B4ITH5 A0A0P8Y9U6 B4PE63 B5RIN9 A0A0K8TRR8 B4IAK9 B3M6T1 A0A1D2NDY4 Q961U7 A0A034VHT6 A0A182WBT0 A0A1L8E1J8 A0A034VM42 W8B513 A0A0K8UVR2 W8B112 A0A1J1IRZ0 A0A0A1XSQ8 A0A0A1WKR5 A0A0A1WH71 A0A3R7PNK7 A0A1B0ABA4 A0A1B6H9E9 W5LY39 W5LY58
PDB
1V9N
E-value=8.54865e-50,
Score=497
Ontologies
GO
PANTHER
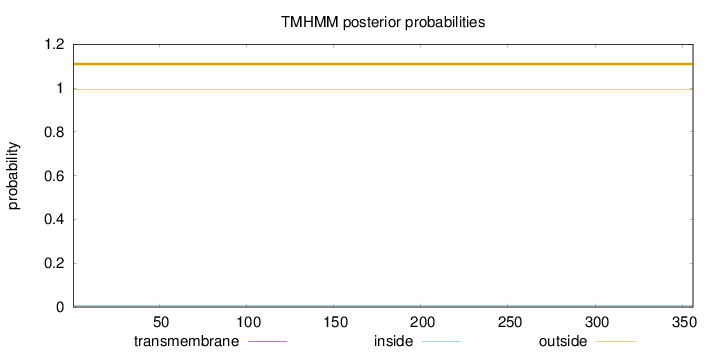
Topology
Length:
356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00247
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00638
outside
1 - 356
Population Genetic Test Statistics
Pi
199.889628
Theta
166.231859
Tajima's D
1.42352
CLR
0.776188
CSRT
0.762361881905905
Interpretation
Uncertain
