Gene
KWMTBOMO11657
Pre Gene Modal
BGIBMGA003990
Annotation
fused_lobes_[Bombyx_mori]
Full name
Beta-hexosaminidase
+ More
Probable beta-hexosaminidase fdl
Probable beta-hexosaminidase fdl
Alternative Name
Protein fused lobes
Location in the cell
Mitochondrial Reliability : 1.129
Sequence
CDS
ATGATGTCGTGGGGTGATGCATTATGGCTTGGCTTGACGGCGAGATTTGCGAGGGTGGGGCGGCTGCGACGAGCCGTGCTCATGCTAGCCGCGGCCGCCTGCACCGCTGCTGCTGTACTCTACTGGAAACAGCAGACTGACGACAGCGCCAACAGACCGCTTCACTCCATGTATTCGGGCATCGAACCTCAGTGGTCTTGGTTATGTCAACACGACCGATGCGAGAGGTATCAAGCATCGGATACGACGACGCTACAGTCGCTGCAGACTTGCAACATGCTGTGCGCCTCCACCCAGCTCTGGCCACAGCCGACGGGGCCTGTCAGCCTCGCATCGGCGGCTGTGCCCGTGAGATCGGACCGCTTCTCCCTCAAAGTTATAGCGTCACCATCACGCGACGTCACCAAACACTTAAACGAAGCGTTTATCGTGATGCAAAATCACATGCGAACTTTAGAACATGGCGTGGTCGGTGAAAACCGTCGATCTGACATAGGACCCCCGCGGGACGTACTGGTGAAGGTATCCGTGAACGGATCCGGCGACCCTCGTATGCGGCTCGACACGAACGAAAGCTACAAACTCGCCTTACGGCCCTCCGGCAATTCCCTCGTCGTAGACATCACAGCGCATTCGTTCTGCGGAGCGAGACACGGCCTCGAAACACTTTTGCAGGTCACCTGGCTGGATCCGTACGCCGGATCGCTACTCATACTAGAAGCGGCAACTGTAGTTGATGCCCCTCGTTTTCCTTACCGTGGCCTGCTTCTGGATACGGCTCGTAATTTTTTCCCCGTCAGTGAGCTTCTCCGCACCATAGACGCGATGGCCGCGAACAAACTAAATACGTTCCACTGGCACGTGAGCGATTCACAATCGTTTCCGTGGAAGTTAGACAGCGCACCCCAACTGGCGCAGCACGGCGCCTACGGACCGGGCGCAGTGTACACGTCCGACGACGTGAGAACAATTGTTAAATACGCACGCATCAGAGGAATCCGGGTGCTGATGGAAATAGACACACCGGCGCACGTCGGTCGAGCTTTCGGTTGGGGTCCGGAGGCTGGCCTCGGCCACTTAGCGCACTGCATAGAGGCGGAACCCTGGAGTTCCTACTGCGGAGAACCTCCGTGCGGCCAACTGAATCCTCGCAACCCTCACATATACGATTTACTAGAACACGTCTACAGGGAGATCATTCAATTAACTGGGGTCGACGACATCTTCCACCTTGGTGGCGATGAGGTGTCGGAACAGTGCTGGGCTAAGCACTTCAATGATACGGATCCGATGGATCTTTGGATGGAGTTCACGCGACAAGCCATGCACGTTCTCGAACGAGCGAATGGGGGTAAAGCGCCAGAGCTCACTCTGCTTTGGTCATCGCGATTGACGCGCTCCCCGTACCTCGAACGCCTCGACCCAAAACGCTTTGGCGTACATGTGTGGGGCGCGTCGCAGTGGCCCGAGTCGCGTGCGGTTTTGGACGCCGGCTTCCGATCGGTGATCTCGCACGTGGACGCCTGGTACCTCGACTGCGGGTTCGGGTCGTGGCGCGACAGCTCGGACGGACACTGCGGGCCGTACCGGTCGTGGCAGCAGGTGTACGAGCACCGACCGTGGGCGACGGAAACGCCCGAGAGCGCGGCATGGCCGGTGGAGGGTGGCGCAGCGTGCCAGTGGACGGAGCAGTTGGGTCCGGGCGGGTTGGACGCGCGCGTGTGGCCCCGGACGGCAGCTCTGGCGGAGCGGCTGTGGGCAGACCGTGCCGAGGGCGCCACGGCGGATGTGTACTTGCGGCTCGACACACAGCGGGCGCGGCTGGTGGCGCGGGGGGTGCGGGCGGCGCCGCTGTGGCCGCGCTGGTGCTCCCACAACCCGCACGCGTGCCTCTAG
Protein
MMSWGDALWLGLTARFARVGRLRRAVLMLAAAACTAAAVLYWKQQTDDSANRPLHSMYSGIEPQWSWLCQHDRCERYQASDTTTLQSLQTCNMLCASTQLWPQPTGPVSLASAAVPVRSDRFSLKVIASPSRDVTKHLNEAFIVMQNHMRTLEHGVVGENRRSDIGPPRDVLVKVSVNGSGDPRMRLDTNESYKLALRPSGNSLVVDITAHSFCGARHGLETLLQVTWLDPYAGSLLILEAATVVDAPRFPYRGLLLDTARNFFPVSELLRTIDAMAANKLNTFHWHVSDSQSFPWKLDSAPQLAQHGAYGPGAVYTSDDVRTIVKYARIRGIRVLMEIDTPAHVGRAFGWGPEAGLGHLAHCIEAEPWSSYCGEPPCGQLNPRNPHIYDLLEHVYREIIQLTGVDDIFHLGGDEVSEQCWAKHFNDTDPMDLWMEFTRQAMHVLERANGGKAPELTLLWSSRLTRSPYLERLDPKRFGVHVWGASQWPESRAVLDAGFRSVISHVDAWYLDCGFGSWRDSSDGHCGPYRSWQQVYEHRPWATETPESAAWPVEGGAACQWTEQLGPGGLDARVWPRTAALAERLWADRAEGATADVYLRLDTQRARLVARGVRAAPLWPRWCSHNPHACL
Summary
Description
Involved in brain restructurization via hormonal control during metamorphosis. Implicated in N-glycan processing.
Catalytic Activity
Hydrolysis of terminal non-reducing N-acetyl-D-hexosamine residues in N-acetyl-beta-D-hexosaminides.
Similarity
Belongs to the glycosyl hydrolase 20 family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Glycoprotein
Glycosidase
Hydrolase
Reference proteome
Signal
Feature
chain Probable beta-hexosaminidase fdl
splice variant In isoform B.
splice variant In isoform B.
Uniprot
D3GGI9
E1CG96
D3GGI8
B1P868
D6PW84
A0A212FNK4
+ More
A0A3S2NQV5 D3GGI7 A0A194QCU8 A0A0L7LFG8 A0A2A4JAM6 A0A0L7KQB6 A0A0G2RLW6 A0A194QYW8 A0A212F886 A0A0L7LKV0 A0A2H1VA55 A5YVX6 A0A194QZT5 A0A2A4JIQ5 A0A194QE79 J3JWB4 U4UHA8 H9J3A9 A0A1Y1MUX2 A0A1W4WN94 A0A1W4WN20 A0A1W4WM34 A0A1W4WC04 A0A0T6BB39 B4H544 Q28X77 A0A0R3NQ30 A0A0R3NWB6 E2AN60 A0A026VXS5 A0A154PQQ9 A0A0R3NVQ4 A0A088ANY0 A0A3L8D4R7 A0A0A7NXB7 A0A195FQK5 A0A3B0JFG0 F4WC87 A0A3B0JKP9 E2BWS6 A0A195C5C4 A0A151XJZ2 Q7QIU7 A0A2A3ES46 A0A1L8E264 A0A195BK61 A0A1J1I7Y3 A0A1J1I989 A0A158N9Y2 A0A1B0CI79 A0A0Q9XGJ7 B4KPZ8 A0A0R1DQK7 B4P590 B4QCX6 A0A0J9RB23 A0A0R1DWG8 A0A1Q3F2V3 A0A0J9RB31 A0A1Q3F2U8 A0A1W4UJA8 B4N602 A0A1W4U6H5 A0A0J9RC20 B0WNY9 A0A0R1DQ79 B4HPK0 A0A0P6IVH7 A0A0L7QN95 A0A232F8H7 K7INU6 A0A0P9AC23 A0A0P8XLY1 A0A0B4LG91 A0A0P8XLW9 Q8WSF3-2 H5V8F4 A0A0B4LF89 Q8WSF3 A0A0Q5W266 A0A0N8NZZ6 A0A0Q5VZG7 B3MEN4 A9UN21 A0A182GY98 B4MDU3 A0A0Q9W0E1 A0A0Q9W9B7 A0A0Q9W007 B3NRZ7 A0A0M4E7X0 A0A084WEE1 A0A336LPK8 A0A336MDZ8
A0A3S2NQV5 D3GGI7 A0A194QCU8 A0A0L7LFG8 A0A2A4JAM6 A0A0L7KQB6 A0A0G2RLW6 A0A194QYW8 A0A212F886 A0A0L7LKV0 A0A2H1VA55 A5YVX6 A0A194QZT5 A0A2A4JIQ5 A0A194QE79 J3JWB4 U4UHA8 H9J3A9 A0A1Y1MUX2 A0A1W4WN94 A0A1W4WN20 A0A1W4WM34 A0A1W4WC04 A0A0T6BB39 B4H544 Q28X77 A0A0R3NQ30 A0A0R3NWB6 E2AN60 A0A026VXS5 A0A154PQQ9 A0A0R3NVQ4 A0A088ANY0 A0A3L8D4R7 A0A0A7NXB7 A0A195FQK5 A0A3B0JFG0 F4WC87 A0A3B0JKP9 E2BWS6 A0A195C5C4 A0A151XJZ2 Q7QIU7 A0A2A3ES46 A0A1L8E264 A0A195BK61 A0A1J1I7Y3 A0A1J1I989 A0A158N9Y2 A0A1B0CI79 A0A0Q9XGJ7 B4KPZ8 A0A0R1DQK7 B4P590 B4QCX6 A0A0J9RB23 A0A0R1DWG8 A0A1Q3F2V3 A0A0J9RB31 A0A1Q3F2U8 A0A1W4UJA8 B4N602 A0A1W4U6H5 A0A0J9RC20 B0WNY9 A0A0R1DQ79 B4HPK0 A0A0P6IVH7 A0A0L7QN95 A0A232F8H7 K7INU6 A0A0P9AC23 A0A0P8XLY1 A0A0B4LG91 A0A0P8XLW9 Q8WSF3-2 H5V8F4 A0A0B4LF89 Q8WSF3 A0A0Q5W266 A0A0N8NZZ6 A0A0Q5VZG7 B3MEN4 A9UN21 A0A182GY98 B4MDU3 A0A0Q9W0E1 A0A0Q9W9B7 A0A0Q9W007 B3NRZ7 A0A0M4E7X0 A0A084WEE1 A0A336LPK8 A0A336MDZ8
EC Number
3.2.1.52
Pubmed
19121390
20547376
18303021
22118469
26354079
26227816
+ More
18342252 18362917 19820115 22516182 23537049 28004739 17994087 15632085 20798317 24508170 30249741 25488985 21719571 12364791 21347285 17550304 22936249 26999592 28648823 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18057021 10623899 12537569 26483478 24438588
18342252 18362917 19820115 22516182 23537049 28004739 17994087 15632085 20798317 24508170 30249741 25488985 21719571 12364791 21347285 17550304 22936249 26999592 28648823 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18057021 10623899 12537569 26483478 24438588
EMBL
BABH01021699
BABH01021700
FJ695481
ACV89847.1
AB540233
BAJ20189.1
+ More
FJ695480 ACV89846.1 EU334747 ODYU01005953 ACA30398.1 SOQ47378.1 GU984637 ADF97235.1 AGBW02005277 OWR55318.1 RSAL01000352 RVE42263.1 FJ695479 ACV89845.1 KQ459185 KPJ03244.1 JTDY01001324 KOB74197.1 NWSH01002205 PCG68909.1 JTDY01007043 KOB65498.1 KP000851 AJG44546.1 KQ460949 KPJ10484.1 AGBW02009781 OWR49928.1 JTDY01000705 KOB76153.1 ODYU01001276 SOQ37272.1 EF592539 KQ971354 ABQ95985.1 EFA06836.2 KPJ10490.1 NWSH01001413 PCG71313.1 KPJ03250.1 BT127532 AEE62494.1 KB632326 ERL92397.1 BABH01021684 GEZM01019981 JAV89483.1 LJIG01002462 KRT84478.1 CH479210 EDW32880.1 CM000071 EAL26439.2 KRT03108.1 KRT03107.1 GL441088 EFN65124.1 KK107796 EZA47639.1 KQ435007 KZC13460.1 KRT03109.1 QOIP01000013 RLU15380.1 KJ786476 AIZ97749.1 KQ981305 KYN42860.1 OUUW01000001 SPP74050.1 GL888070 EGI68067.1 SPP74049.1 GL451188 EFN79790.1 KQ978292 KYM95386.1 KQ982052 KYQ60598.1 AAAB01008807 EAA03943.4 KZ288193 PBC34036.1 GFDF01001338 JAV12746.1 KQ976455 KYM85038.1 CVRI01000043 CRK96299.1 CRK96298.1 ADTU01009908 AJWK01013083 CH933808 KRG03902.1 EDW08100.1 CM000158 KRJ99444.1 EDW90752.2 CM000362 CM002911 EDX06787.1 KMY93230.1 KMY93226.1 KRJ99443.1 GFDL01013164 JAV21881.1 KMY93227.1 GFDL01013165 JAV21880.1 CH964154 EDW79791.2 KMY93229.1 DS232017 EDS32008.1 KRJ99445.1 CH480816 EDW47584.1 GDUN01000092 JAN95827.1 KQ414860 KOC60039.1 NNAY01000660 OXU27126.1 CH902619 KPU75705.1 KPU75706.1 AE013599 AHN56149.1 KPU75704.1 AF323977 AY113418 AY061037 AAL28585.1 AAL55992.1 AAM29423.1 BT133255 AFC17499.1 AHN56148.1 CH954179 KQS62831.1 KPU75708.1 KQS62830.1 EDV35498.2 BT031185 ABY20426.1 JXUM01097258 JXUM01097259 JXUM01097260 JXUM01097261 CH940662 EDW58708.1 KRF78418.1 KRF78417.1 KRF78419.1 EDV56299.2 CP012524 ALC40696.1 ATLV01023205 KE525341 KFB48585.1 UFQS01000102 UFQT01000102 SSW99622.1 SSX20002.1 UFQT01001050 SSX28632.1
FJ695480 ACV89846.1 EU334747 ODYU01005953 ACA30398.1 SOQ47378.1 GU984637 ADF97235.1 AGBW02005277 OWR55318.1 RSAL01000352 RVE42263.1 FJ695479 ACV89845.1 KQ459185 KPJ03244.1 JTDY01001324 KOB74197.1 NWSH01002205 PCG68909.1 JTDY01007043 KOB65498.1 KP000851 AJG44546.1 KQ460949 KPJ10484.1 AGBW02009781 OWR49928.1 JTDY01000705 KOB76153.1 ODYU01001276 SOQ37272.1 EF592539 KQ971354 ABQ95985.1 EFA06836.2 KPJ10490.1 NWSH01001413 PCG71313.1 KPJ03250.1 BT127532 AEE62494.1 KB632326 ERL92397.1 BABH01021684 GEZM01019981 JAV89483.1 LJIG01002462 KRT84478.1 CH479210 EDW32880.1 CM000071 EAL26439.2 KRT03108.1 KRT03107.1 GL441088 EFN65124.1 KK107796 EZA47639.1 KQ435007 KZC13460.1 KRT03109.1 QOIP01000013 RLU15380.1 KJ786476 AIZ97749.1 KQ981305 KYN42860.1 OUUW01000001 SPP74050.1 GL888070 EGI68067.1 SPP74049.1 GL451188 EFN79790.1 KQ978292 KYM95386.1 KQ982052 KYQ60598.1 AAAB01008807 EAA03943.4 KZ288193 PBC34036.1 GFDF01001338 JAV12746.1 KQ976455 KYM85038.1 CVRI01000043 CRK96299.1 CRK96298.1 ADTU01009908 AJWK01013083 CH933808 KRG03902.1 EDW08100.1 CM000158 KRJ99444.1 EDW90752.2 CM000362 CM002911 EDX06787.1 KMY93230.1 KMY93226.1 KRJ99443.1 GFDL01013164 JAV21881.1 KMY93227.1 GFDL01013165 JAV21880.1 CH964154 EDW79791.2 KMY93229.1 DS232017 EDS32008.1 KRJ99445.1 CH480816 EDW47584.1 GDUN01000092 JAN95827.1 KQ414860 KOC60039.1 NNAY01000660 OXU27126.1 CH902619 KPU75705.1 KPU75706.1 AE013599 AHN56149.1 KPU75704.1 AF323977 AY113418 AY061037 AAL28585.1 AAL55992.1 AAM29423.1 BT133255 AFC17499.1 AHN56148.1 CH954179 KQS62831.1 KPU75708.1 KQS62830.1 EDV35498.2 BT031185 ABY20426.1 JXUM01097258 JXUM01097259 JXUM01097260 JXUM01097261 CH940662 EDW58708.1 KRF78418.1 KRF78417.1 KRF78419.1 EDV56299.2 CP012524 ALC40696.1 ATLV01023205 KE525341 KFB48585.1 UFQS01000102 UFQT01000102 SSW99622.1 SSX20002.1 UFQT01001050 SSX28632.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053268
UP000037510
UP000218220
+ More
UP000053240 UP000007266 UP000030742 UP000192223 UP000008744 UP000001819 UP000000311 UP000053097 UP000076502 UP000005203 UP000279307 UP000078541 UP000268350 UP000007755 UP000008237 UP000078542 UP000075809 UP000007062 UP000242457 UP000078540 UP000183832 UP000005205 UP000092461 UP000009192 UP000002282 UP000000304 UP000192221 UP000007798 UP000002320 UP000001292 UP000053825 UP000215335 UP000002358 UP000007801 UP000000803 UP000008711 UP000069940 UP000008792 UP000092553 UP000030765
UP000053240 UP000007266 UP000030742 UP000192223 UP000008744 UP000001819 UP000000311 UP000053097 UP000076502 UP000005203 UP000279307 UP000078541 UP000268350 UP000007755 UP000008237 UP000078542 UP000075809 UP000007062 UP000242457 UP000078540 UP000183832 UP000005205 UP000092461 UP000009192 UP000002282 UP000000304 UP000192221 UP000007798 UP000002320 UP000001292 UP000053825 UP000215335 UP000002358 UP000007801 UP000000803 UP000008711 UP000069940 UP000008792 UP000092553 UP000030765
Interpro
Gene 3D
ProteinModelPortal
D3GGI9
E1CG96
D3GGI8
B1P868
D6PW84
A0A212FNK4
+ More
A0A3S2NQV5 D3GGI7 A0A194QCU8 A0A0L7LFG8 A0A2A4JAM6 A0A0L7KQB6 A0A0G2RLW6 A0A194QYW8 A0A212F886 A0A0L7LKV0 A0A2H1VA55 A5YVX6 A0A194QZT5 A0A2A4JIQ5 A0A194QE79 J3JWB4 U4UHA8 H9J3A9 A0A1Y1MUX2 A0A1W4WN94 A0A1W4WN20 A0A1W4WM34 A0A1W4WC04 A0A0T6BB39 B4H544 Q28X77 A0A0R3NQ30 A0A0R3NWB6 E2AN60 A0A026VXS5 A0A154PQQ9 A0A0R3NVQ4 A0A088ANY0 A0A3L8D4R7 A0A0A7NXB7 A0A195FQK5 A0A3B0JFG0 F4WC87 A0A3B0JKP9 E2BWS6 A0A195C5C4 A0A151XJZ2 Q7QIU7 A0A2A3ES46 A0A1L8E264 A0A195BK61 A0A1J1I7Y3 A0A1J1I989 A0A158N9Y2 A0A1B0CI79 A0A0Q9XGJ7 B4KPZ8 A0A0R1DQK7 B4P590 B4QCX6 A0A0J9RB23 A0A0R1DWG8 A0A1Q3F2V3 A0A0J9RB31 A0A1Q3F2U8 A0A1W4UJA8 B4N602 A0A1W4U6H5 A0A0J9RC20 B0WNY9 A0A0R1DQ79 B4HPK0 A0A0P6IVH7 A0A0L7QN95 A0A232F8H7 K7INU6 A0A0P9AC23 A0A0P8XLY1 A0A0B4LG91 A0A0P8XLW9 Q8WSF3-2 H5V8F4 A0A0B4LF89 Q8WSF3 A0A0Q5W266 A0A0N8NZZ6 A0A0Q5VZG7 B3MEN4 A9UN21 A0A182GY98 B4MDU3 A0A0Q9W0E1 A0A0Q9W9B7 A0A0Q9W007 B3NRZ7 A0A0M4E7X0 A0A084WEE1 A0A336LPK8 A0A336MDZ8
A0A3S2NQV5 D3GGI7 A0A194QCU8 A0A0L7LFG8 A0A2A4JAM6 A0A0L7KQB6 A0A0G2RLW6 A0A194QYW8 A0A212F886 A0A0L7LKV0 A0A2H1VA55 A5YVX6 A0A194QZT5 A0A2A4JIQ5 A0A194QE79 J3JWB4 U4UHA8 H9J3A9 A0A1Y1MUX2 A0A1W4WN94 A0A1W4WN20 A0A1W4WM34 A0A1W4WC04 A0A0T6BB39 B4H544 Q28X77 A0A0R3NQ30 A0A0R3NWB6 E2AN60 A0A026VXS5 A0A154PQQ9 A0A0R3NVQ4 A0A088ANY0 A0A3L8D4R7 A0A0A7NXB7 A0A195FQK5 A0A3B0JFG0 F4WC87 A0A3B0JKP9 E2BWS6 A0A195C5C4 A0A151XJZ2 Q7QIU7 A0A2A3ES46 A0A1L8E264 A0A195BK61 A0A1J1I7Y3 A0A1J1I989 A0A158N9Y2 A0A1B0CI79 A0A0Q9XGJ7 B4KPZ8 A0A0R1DQK7 B4P590 B4QCX6 A0A0J9RB23 A0A0R1DWG8 A0A1Q3F2V3 A0A0J9RB31 A0A1Q3F2U8 A0A1W4UJA8 B4N602 A0A1W4U6H5 A0A0J9RC20 B0WNY9 A0A0R1DQ79 B4HPK0 A0A0P6IVH7 A0A0L7QN95 A0A232F8H7 K7INU6 A0A0P9AC23 A0A0P8XLY1 A0A0B4LG91 A0A0P8XLW9 Q8WSF3-2 H5V8F4 A0A0B4LF89 Q8WSF3 A0A0Q5W266 A0A0N8NZZ6 A0A0Q5VZG7 B3MEN4 A9UN21 A0A182GY98 B4MDU3 A0A0Q9W0E1 A0A0Q9W9B7 A0A0Q9W007 B3NRZ7 A0A0M4E7X0 A0A084WEE1 A0A336LPK8 A0A336MDZ8
PDB
3S6T
E-value=7.64911e-95,
Score=888
Ontologies
PATHWAY
GO
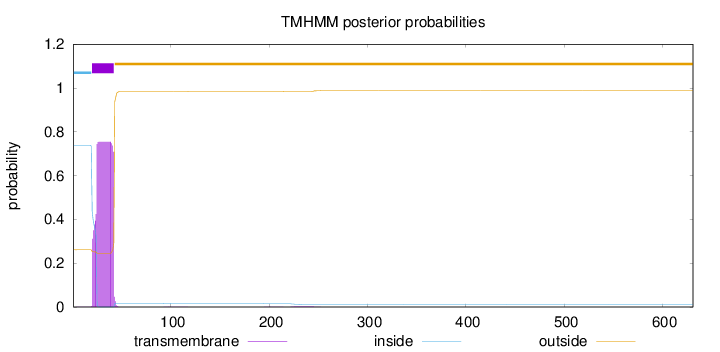
Topology
Length:
631
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.533
Exp number, first 60 AAs:
15.44789
Total prob of N-in:
0.73885
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 631
Population Genetic Test Statistics
Pi
222.117702
Theta
166.859805
Tajima's D
0.830436
CLR
0.31086
CSRT
0.611669416529174
Interpretation
Uncertain
