Pre Gene Modal
BGIBMGA003991
Annotation
PREDICTED:_bis(5'-nucleosyl)-tetraphosphatase_[asymmetrical]_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.718 Extracellular Reliability : 1.782
Sequence
CDS
ATGTCTTCTTTAAGAGCCGCAGGTTTGGTTATTTTCAGAAATTATAATCAAATTATACAATTCCTATTACTGCAAACATCCTACGGAGAACATCATTGGACACCACCAAAAGGACATGTTGATCCTGGAGAAACTGATTGGATGACAGCTTTGAGAGAAACAAAAGAAGAAGCCGGTCTGTGCGAAGATCACTTAGATATTTACAAAGACATAAACAAAACTTTAAATTATGAAGTAAACGGAGAACCCAAAACTGTTGTGTACTGGTTGGCAAAGCTTAAAAATCCTGAACAGACAGTCACCCTTTCATCTGAACACCAAGATATGAAGTGGCTCTCATTGCAAGAAGCACAGGAAATATCAAAATACGAAGACATGAGACAATTGCTTGCGGAATTTTATGAGAAATGTAAAAGTAGATAA
Protein
MSSLRAAGLVIFRNYNQIIQFLLLQTSYGEHHWTPPKGHVDPGETDWMTALRETKEEAGLCEDHLDIYKDINKTLNYEVNGEPKTVVYWLAKLKNPEQTVTLSSEHQDMKWLSLQEAQEISKYEDMRQLLAEFYEKCKSR
Summary
Uniprot
H9J3A4
S4PD63
A0A2A4JIT2
A0A212FNI8
A0A2H1WF52
A0A194R3R7
+ More
A0A194QIH0 I4DK53 A0A026WTK2 E2AVU4 A0A183FQ35 A0A3P7Y451 V5GNZ0 A0A2A3EP27 A0A088AUA2 E2BKT2 C3ZGP8 A0A0N4YUF8 A0A310S4T4 J3JX45 A0A1B6G3I1 A0A0M9A2Z5 A0A1B6LV56 A0A182UKP7 F4W7Z4 A0A182VC87 A0A195D7T9 A0A151X4L3 A0A182MMY0 A0A195C731 A0A182X162 A0A182IBW6 A0A1Y1NIU1 A0A182WHH8 A0A182RRJ4 Q7Q4N2 A0A0A1X4V1 A0A182NUW0 A0A182P3B4 A0A0P4Y2W4 A0A0P5K1C6 A0A195EZC9 A0A182Y5K1 A0A182QT00 A0A1I8QBH2 K1QKT4 A0A0K8V9Y5 A0A2G9UNP2 A0A0N4X1C6 H3ELP0 A0A182JNH0 A0A182JAS2 A0A3Q3CCD9 A0A139WFV7 A0A3P9Q9Y3 A0A016UV17 A0A084VL62 A0A210QDU6 W8C4U7 B0W8U4 A0A3Q2G0R0 A0A1Q3FFY0 A0A016UUD2 A0A146P4U5 A5D8R0 A0A0K8TS11 A0A0P5FGN7 A0A0B8RWT4 A0A195BR44 A0A158NDT8 A0A232F260 A0A2M4AXG2 A0A2M4AXQ2 A0A2I4BXP2 W8BQE8 A0A3B3XC97 A0A087XWL8 A0A3B3UA41 A0A0L0BVC7 A0A0B7A9P8 A0A2D4IIM1 M3ZMI0 A0A3B5LNA5 Q6DG97 A0A1A7Y9M2 V8NLQ4 A0A2D4L8Q5 A0A0C9QPN9 A0A3P8PME7 A0A3P9DP38 A0A2M3ZBU7 A0A2M4C2L3 A0A1L8E8U4 A0A0L7RAW6 A0A2A6BLR8 A0A1S3P613 A0A1S3P606 W8BIZ0 A0A1S3P609 A0A1A9XMQ2
A0A194QIH0 I4DK53 A0A026WTK2 E2AVU4 A0A183FQ35 A0A3P7Y451 V5GNZ0 A0A2A3EP27 A0A088AUA2 E2BKT2 C3ZGP8 A0A0N4YUF8 A0A310S4T4 J3JX45 A0A1B6G3I1 A0A0M9A2Z5 A0A1B6LV56 A0A182UKP7 F4W7Z4 A0A182VC87 A0A195D7T9 A0A151X4L3 A0A182MMY0 A0A195C731 A0A182X162 A0A182IBW6 A0A1Y1NIU1 A0A182WHH8 A0A182RRJ4 Q7Q4N2 A0A0A1X4V1 A0A182NUW0 A0A182P3B4 A0A0P4Y2W4 A0A0P5K1C6 A0A195EZC9 A0A182Y5K1 A0A182QT00 A0A1I8QBH2 K1QKT4 A0A0K8V9Y5 A0A2G9UNP2 A0A0N4X1C6 H3ELP0 A0A182JNH0 A0A182JAS2 A0A3Q3CCD9 A0A139WFV7 A0A3P9Q9Y3 A0A016UV17 A0A084VL62 A0A210QDU6 W8C4U7 B0W8U4 A0A3Q2G0R0 A0A1Q3FFY0 A0A016UUD2 A0A146P4U5 A5D8R0 A0A0K8TS11 A0A0P5FGN7 A0A0B8RWT4 A0A195BR44 A0A158NDT8 A0A232F260 A0A2M4AXG2 A0A2M4AXQ2 A0A2I4BXP2 W8BQE8 A0A3B3XC97 A0A087XWL8 A0A3B3UA41 A0A0L0BVC7 A0A0B7A9P8 A0A2D4IIM1 M3ZMI0 A0A3B5LNA5 Q6DG97 A0A1A7Y9M2 V8NLQ4 A0A2D4L8Q5 A0A0C9QPN9 A0A3P8PME7 A0A3P9DP38 A0A2M3ZBU7 A0A2M4C2L3 A0A1L8E8U4 A0A0L7RAW6 A0A2A6BLR8 A0A1S3P613 A0A1S3P606 W8BIZ0 A0A1S3P609 A0A1A9XMQ2
Pubmed
19121390
23622113
22118469
26354079
22651552
24508170
+ More
30249741 20798317 18563158 22516182 23537049 21719571 28004739 12364791 14747013 17210077 25830018 25244985 22992520 18806794 18362917 19820115 25730766 24438588 28812685 24495485 26369729 25476704 21347285 28648823 26108605 23542700 23594743 24297900 25186727
30249741 20798317 18563158 22516182 23537049 21719571 28004739 12364791 14747013 17210077 25830018 25244985 22992520 18806794 18362917 19820115 25730766 24438588 28812685 24495485 26369729 25476704 21347285 28648823 26108605 23542700 23594743 24297900 25186727
EMBL
BABH01021699
GAIX01003786
JAA88774.1
NWSH01001413
PCG71320.1
AGBW02005277
+ More
OWR55316.1 ODYU01008187 SOQ51576.1 KQ460949 KPJ10486.1 KQ459185 KPJ03246.1 AK401671 BAM18293.1 KK107126 QOIP01000008 EZA58429.1 RLU19254.1 GL443213 EFN62366.1 UZAH01026555 VDO82356.1 GALX01006468 JAB61998.1 KZ288203 PBC33450.1 GL448826 EFN83681.1 GG666620 EEN48274.1 UYSL01025628 VDL84619.1 KQ775154 OAD52275.1 APGK01008348 APGK01013783 APGK01057784 BT127813 KB741282 KB739113 KB737669 KB631802 AEE62775.1 ENN70778.1 ENN82381.1 ENN82966.1 ERL86228.1 GECZ01012771 JAS56998.1 KQ435752 KOX76220.1 GEBQ01012416 JAT27561.1 GL887888 EGI69664.1 KQ981153 KYN08922.1 KQ982548 KYQ55229.1 AXCM01001953 KQ978251 KYM95981.1 APCN01000751 GEZM01003452 JAV96800.1 AAAB01008964 EAA12242.4 GBXI01008579 JAD05713.1 GDIP01242684 LRGB01002101 JAI80717.1 KZS09164.1 GDIQ01191182 JAK60543.1 KQ981906 KYN33229.1 AXCN02000748 JH818171 EKC37377.1 GDHF01016565 JAI35749.1 KZ345984 PIO71332.1 UZAF01020339 VDO68993.1 KQ971351 KYB26853.1 KE124844 JARK01001362 EPB77135.1 EYC18821.1 ATLV01014417 KE524970 KFB38706.1 NEDP02004063 OWF46912.1 GAMC01005274 JAC01282.1 DS231860 EDS39323.1 GFDL01008633 JAV26412.1 EYC18820.1 GCES01147341 JAQ38981.1 BC141775 AAI41776.1 GDAI01000665 JAI16938.1 GDIQ01257700 JAJ94024.1 GBSH01002714 JAG66313.1 KQ976424 KYM88431.1 ADTU01012968 NNAY01001180 OXU24834.1 GGFK01012165 MBW45486.1 GGFK01012204 MBW45525.1 GAMC01005273 JAC01283.1 AYCK01005653 JRES01001284 JRES01000418 KNC23938.1 KNC31446.1 HACG01030578 CEK77443.1 IACK01104237 LAA84046.1 CU694487 BC076454 AAH76454.1 HADW01012119 HADX01004729 SBP26961.1 AZIM01003163 ETE62598.1 IACL01123176 LAB17349.1 GBYB01005589 JAG75356.1 GGFM01005273 MBW26024.1 GGFJ01010411 MBW59552.1 GFDG01003658 JAV15141.1 KQ414617 KOC68062.1 ABKE03000093 PDM66847.1 GAMC01005275 JAC01281.1
OWR55316.1 ODYU01008187 SOQ51576.1 KQ460949 KPJ10486.1 KQ459185 KPJ03246.1 AK401671 BAM18293.1 KK107126 QOIP01000008 EZA58429.1 RLU19254.1 GL443213 EFN62366.1 UZAH01026555 VDO82356.1 GALX01006468 JAB61998.1 KZ288203 PBC33450.1 GL448826 EFN83681.1 GG666620 EEN48274.1 UYSL01025628 VDL84619.1 KQ775154 OAD52275.1 APGK01008348 APGK01013783 APGK01057784 BT127813 KB741282 KB739113 KB737669 KB631802 AEE62775.1 ENN70778.1 ENN82381.1 ENN82966.1 ERL86228.1 GECZ01012771 JAS56998.1 KQ435752 KOX76220.1 GEBQ01012416 JAT27561.1 GL887888 EGI69664.1 KQ981153 KYN08922.1 KQ982548 KYQ55229.1 AXCM01001953 KQ978251 KYM95981.1 APCN01000751 GEZM01003452 JAV96800.1 AAAB01008964 EAA12242.4 GBXI01008579 JAD05713.1 GDIP01242684 LRGB01002101 JAI80717.1 KZS09164.1 GDIQ01191182 JAK60543.1 KQ981906 KYN33229.1 AXCN02000748 JH818171 EKC37377.1 GDHF01016565 JAI35749.1 KZ345984 PIO71332.1 UZAF01020339 VDO68993.1 KQ971351 KYB26853.1 KE124844 JARK01001362 EPB77135.1 EYC18821.1 ATLV01014417 KE524970 KFB38706.1 NEDP02004063 OWF46912.1 GAMC01005274 JAC01282.1 DS231860 EDS39323.1 GFDL01008633 JAV26412.1 EYC18820.1 GCES01147341 JAQ38981.1 BC141775 AAI41776.1 GDAI01000665 JAI16938.1 GDIQ01257700 JAJ94024.1 GBSH01002714 JAG66313.1 KQ976424 KYM88431.1 ADTU01012968 NNAY01001180 OXU24834.1 GGFK01012165 MBW45486.1 GGFK01012204 MBW45525.1 GAMC01005273 JAC01283.1 AYCK01005653 JRES01001284 JRES01000418 KNC23938.1 KNC31446.1 HACG01030578 CEK77443.1 IACK01104237 LAA84046.1 CU694487 BC076454 AAH76454.1 HADW01012119 HADX01004729 SBP26961.1 AZIM01003163 ETE62598.1 IACL01123176 LAB17349.1 GBYB01005589 JAG75356.1 GGFM01005273 MBW26024.1 GGFJ01010411 MBW59552.1 GFDG01003658 JAV15141.1 KQ414617 KOC68062.1 ABKE03000093 PDM66847.1 GAMC01005275 JAC01281.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000053097
+ More
UP000279307 UP000000311 UP000050761 UP000242457 UP000005203 UP000008237 UP000001554 UP000038043 UP000271162 UP000019118 UP000030742 UP000053105 UP000075902 UP000007755 UP000075903 UP000078492 UP000075809 UP000075883 UP000078542 UP000076407 UP000075840 UP000075920 UP000075900 UP000007062 UP000075884 UP000075885 UP000076858 UP000078541 UP000076408 UP000075886 UP000095300 UP000005408 UP000038042 UP000268014 UP000005239 UP000075881 UP000075880 UP000264840 UP000007266 UP000242638 UP000024635 UP000030765 UP000242188 UP000002320 UP000265020 UP000265000 UP000078540 UP000005205 UP000215335 UP000192220 UP000261480 UP000028760 UP000261500 UP000037069 UP000002852 UP000261380 UP000000437 UP000265100 UP000265160 UP000053825 UP000232936 UP000087266 UP000092443
UP000279307 UP000000311 UP000050761 UP000242457 UP000005203 UP000008237 UP000001554 UP000038043 UP000271162 UP000019118 UP000030742 UP000053105 UP000075902 UP000007755 UP000075903 UP000078492 UP000075809 UP000075883 UP000078542 UP000076407 UP000075840 UP000075920 UP000075900 UP000007062 UP000075884 UP000075885 UP000076858 UP000078541 UP000076408 UP000075886 UP000095300 UP000005408 UP000038042 UP000268014 UP000005239 UP000075881 UP000075880 UP000264840 UP000007266 UP000242638 UP000024635 UP000030765 UP000242188 UP000002320 UP000265020 UP000265000 UP000078540 UP000005205 UP000215335 UP000192220 UP000261480 UP000028760 UP000261500 UP000037069 UP000002852 UP000261380 UP000000437 UP000265100 UP000265160 UP000053825 UP000232936 UP000087266 UP000092443
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J3A4
S4PD63
A0A2A4JIT2
A0A212FNI8
A0A2H1WF52
A0A194R3R7
+ More
A0A194QIH0 I4DK53 A0A026WTK2 E2AVU4 A0A183FQ35 A0A3P7Y451 V5GNZ0 A0A2A3EP27 A0A088AUA2 E2BKT2 C3ZGP8 A0A0N4YUF8 A0A310S4T4 J3JX45 A0A1B6G3I1 A0A0M9A2Z5 A0A1B6LV56 A0A182UKP7 F4W7Z4 A0A182VC87 A0A195D7T9 A0A151X4L3 A0A182MMY0 A0A195C731 A0A182X162 A0A182IBW6 A0A1Y1NIU1 A0A182WHH8 A0A182RRJ4 Q7Q4N2 A0A0A1X4V1 A0A182NUW0 A0A182P3B4 A0A0P4Y2W4 A0A0P5K1C6 A0A195EZC9 A0A182Y5K1 A0A182QT00 A0A1I8QBH2 K1QKT4 A0A0K8V9Y5 A0A2G9UNP2 A0A0N4X1C6 H3ELP0 A0A182JNH0 A0A182JAS2 A0A3Q3CCD9 A0A139WFV7 A0A3P9Q9Y3 A0A016UV17 A0A084VL62 A0A210QDU6 W8C4U7 B0W8U4 A0A3Q2G0R0 A0A1Q3FFY0 A0A016UUD2 A0A146P4U5 A5D8R0 A0A0K8TS11 A0A0P5FGN7 A0A0B8RWT4 A0A195BR44 A0A158NDT8 A0A232F260 A0A2M4AXG2 A0A2M4AXQ2 A0A2I4BXP2 W8BQE8 A0A3B3XC97 A0A087XWL8 A0A3B3UA41 A0A0L0BVC7 A0A0B7A9P8 A0A2D4IIM1 M3ZMI0 A0A3B5LNA5 Q6DG97 A0A1A7Y9M2 V8NLQ4 A0A2D4L8Q5 A0A0C9QPN9 A0A3P8PME7 A0A3P9DP38 A0A2M3ZBU7 A0A2M4C2L3 A0A1L8E8U4 A0A0L7RAW6 A0A2A6BLR8 A0A1S3P613 A0A1S3P606 W8BIZ0 A0A1S3P609 A0A1A9XMQ2
A0A194QIH0 I4DK53 A0A026WTK2 E2AVU4 A0A183FQ35 A0A3P7Y451 V5GNZ0 A0A2A3EP27 A0A088AUA2 E2BKT2 C3ZGP8 A0A0N4YUF8 A0A310S4T4 J3JX45 A0A1B6G3I1 A0A0M9A2Z5 A0A1B6LV56 A0A182UKP7 F4W7Z4 A0A182VC87 A0A195D7T9 A0A151X4L3 A0A182MMY0 A0A195C731 A0A182X162 A0A182IBW6 A0A1Y1NIU1 A0A182WHH8 A0A182RRJ4 Q7Q4N2 A0A0A1X4V1 A0A182NUW0 A0A182P3B4 A0A0P4Y2W4 A0A0P5K1C6 A0A195EZC9 A0A182Y5K1 A0A182QT00 A0A1I8QBH2 K1QKT4 A0A0K8V9Y5 A0A2G9UNP2 A0A0N4X1C6 H3ELP0 A0A182JNH0 A0A182JAS2 A0A3Q3CCD9 A0A139WFV7 A0A3P9Q9Y3 A0A016UV17 A0A084VL62 A0A210QDU6 W8C4U7 B0W8U4 A0A3Q2G0R0 A0A1Q3FFY0 A0A016UUD2 A0A146P4U5 A5D8R0 A0A0K8TS11 A0A0P5FGN7 A0A0B8RWT4 A0A195BR44 A0A158NDT8 A0A232F260 A0A2M4AXG2 A0A2M4AXQ2 A0A2I4BXP2 W8BQE8 A0A3B3XC97 A0A087XWL8 A0A3B3UA41 A0A0L0BVC7 A0A0B7A9P8 A0A2D4IIM1 M3ZMI0 A0A3B5LNA5 Q6DG97 A0A1A7Y9M2 V8NLQ4 A0A2D4L8Q5 A0A0C9QPN9 A0A3P8PME7 A0A3P9DP38 A0A2M3ZBU7 A0A2M4C2L3 A0A1L8E8U4 A0A0L7RAW6 A0A2A6BLR8 A0A1S3P613 A0A1S3P606 W8BIZ0 A0A1S3P609 A0A1A9XMQ2
PDB
1KTG
E-value=1.10421e-35,
Score=369
Ontologies
PATHWAY
GO
Topology
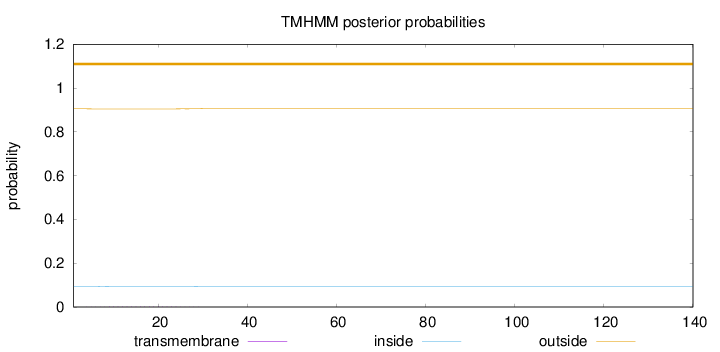
Length:
140
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03254
Exp number, first 60 AAs:
0.03254
Total prob of N-in:
0.09500
outside
1 - 140
Population Genetic Test Statistics
Pi
128.118015
Theta
130.493233
Tajima's D
-0.254542
CLR
63.277526
CSRT
0.306734663266837
Interpretation
Uncertain
