Pre Gene Modal
BGIBMGA004000
Annotation
alpha-N-acetylgalactosaminidase_precursor_[Bombyx_mori]
Full name
Alpha-galactosidase
Location in the cell
Cytoplasmic Reliability : 2.502
Sequence
CDS
ATGATAAGGTTGATTTTAGTTTTTTTAACAATATCGCACAGCTACGCCTTGAATAATGGATTAGCAATGACACCACCAATGGGCTGGCTGACATGGGAACGTTTTAGGTGTATAACCGACTGCGAGAAATATCCTAACGAATGCATAAGCGAGAGCCTAATAAAAAGGACAGCAGATTTGATGGTCAGCGAAGGATATCTAGATGCAGGATATGAATATTTAGGTATCGACGATTGTTGGTTAGAGAAGACACGCGATGATAAAGGTCGGCTGGTACCCGACAGGAAACGATTTCCAAATGGGATGAAGGCCATAGCTGATTATATCCACAGCAAAGGCTTAAAGTTCGGCATGTACGAAGATTATGGAAACCTCACATGTGCCGGTTACCCCGGGGTGCTTGGAAACGAAAAAATCGATATCAACACATTTGTCGAATGGGAAATCGATTATTTGAAGCTCGATGGATGTTATATTGACCCGATACAAATGGATAAAGGCTATCCAGACTTCGGTAAATTGTTGAACGCTACTGGACGATCCATCTTGTACTCCTGTAGCTGGCCAGCATATCAAGAGGAAAAAAAGATATTGCCAAACTATGCATCGATAGCAGAGCACTGTAACCTTTGGCGCAATTATGATGATATAGAAGACTCTTGGGCATCTCTTACGAAGATCATGGCCTGGTTTGGTGACAACCAGGACAGGCTTGCTCAGCACGCCGGGCCTGGACACTGGAACGACCCTGATATGTTACTAATTGGCAACTTCGGTCTATCGGTGGACCAAGCTAAAGTACAAATGGCTGTTTGGGCGATCCTAGCAGCACCGCTTCTGATCAGTACTGATTTGGCAACGATAAGGCCAGAATTCAAACAAATATTACTGAACAGAGACATTATAGCCGTCAACCAAGACAAATTGGGTAAACAAGGATTGCGTGTTTGGAAAAAAATGGGATCTGGATCCAGGTCCAAATTAGAGATATGGAAAAGAGAACTTTATGACGGTTCCTTTGCTATGGCATTTGTGAGTTATCGGGACGATGGAATACCTTACGCAGCCAAATTTACGTATTATGACATGCAATTACCTCAGTCGGAGTTTGAAGTACAGGACTTGTACAAAGAAGAAGAAGTTCGGTCATGGGATGGCACAAGCGACTTCGAAGTTCGGATTAACCCTTCTGGAGTTAAATTTTACAAATTCATTAGCAAAAAATCTATGTCGGAGATTTTGGATATAACAGTAAAAATGATTGTGAAGAATCAAGTGTGTAATTAG
Protein
MIRLILVFLTISHSYALNNGLAMTPPMGWLTWERFRCITDCEKYPNECISESLIKRTADLMVSEGYLDAGYEYLGIDDCWLEKTRDDKGRLVPDRKRFPNGMKAIADYIHSKGLKFGMYEDYGNLTCAGYPGVLGNEKIDINTFVEWEIDYLKLDGCYIDPIQMDKGYPDFGKLLNATGRSILYSCSWPAYQEEKKILPNYASIAEHCNLWRNYDDIEDSWASLTKIMAWFGDNQDRLAQHAGPGHWNDPDMLLIGNFGLSVDQAKVQMAVWAILAAPLLISTDLATIRPEFKQILLNRDIIAVNQDKLGKQGLRVWKKMGSGSRSKLEIWKRELYDGSFAMAFVSYRDDGIPYAAKFTYYDMQLPQSEFEVQDLYKEEEVRSWDGTSDFEVRINPSGVKFYKFISKKSMSEILDITVKMIVKNQVCN
Summary
Subunit
Homodimer.
Similarity
Belongs to the glycosyl hydrolase 27 family.
Feature
chain Alpha-galactosidase
Uniprot
Q2F5Z9
A0A2H1VKD1
A0A2A4JM48
A0A212FA86
A0A194QIH9
A0A2A4JM49
+ More
A0A2A4JKI6 A0A1I8N189 Q7K127 B4HSW9 B3NPJ7 B4QHX7 B4P682 A0A1I8PUH1 W8B716 A0A1W4UXD8 A0A034VJ62 A0A0L0C4I6 A0A0K8WDV6 A0A0A1X480 B3MFN5 Q291S9 D1ZZP5 A0A1B0AF32 B4MPF1 A0A0K8TAY4 B4MFP7 A0A3B0IZ83 A0A1B6FU31 A0A067RL82 A0A2J7Q6R5 Q17A03 A0A1L8DR08 A0A182Q255 A0A1L8DQP1 A0A2P8ZLY1 A0A182IPD0 B4J8F0 A0A1Q3G451 A0A1L8DQU5 A0A182MV14 A0A182VNL1 T1PFY5 A0A1L8DQP5 A0A1Q3G433 A0A182Y8D8 B4GAB8 A0A182L619 Q7Q1V0 A0A2S2QTQ5 A0A224XQS0 A0A182ICP7 A0A084VNI9 A0A2M4BML2 T1HM73 A0A2M4AC36 A0A2M3Z177 A0A2M4ACK0 A0A2M4DMQ3 A0A2M4DMN4 A0A1A9X4X6 A0A2M4DMS1 A0A1B6M6K3 A0A1A9XWH4 A0A1J1IRP1 B3MPM5 A0A151WST7 A0A195BWB5 A0A195CCG6 A0A026WYH3 J9K999 B4M8H2 A0A158NE94 A0A1W4UUS4 B4KRT1 B4NZ59 Q9VL27 J3JUA1 A0A336M9C7 Q8MYY3 A0A195FQA7 B3N935 J9JRF8 B4JP37 B4HWH1 B4Q8Q4 A0A1J1HJ44 F4WJD6 A0A1A9VU43 A0A1B0FRD5 A0A2S2ND41 A0A1A9UT81 A0A1A9ZVN6 T1HJA0 A0A151J0R5 A0A1Y1M843 A0A0L7QQI1 U4UPP5 B4G7B6 A0A1A9WLA3
A0A2A4JKI6 A0A1I8N189 Q7K127 B4HSW9 B3NPJ7 B4QHX7 B4P682 A0A1I8PUH1 W8B716 A0A1W4UXD8 A0A034VJ62 A0A0L0C4I6 A0A0K8WDV6 A0A0A1X480 B3MFN5 Q291S9 D1ZZP5 A0A1B0AF32 B4MPF1 A0A0K8TAY4 B4MFP7 A0A3B0IZ83 A0A1B6FU31 A0A067RL82 A0A2J7Q6R5 Q17A03 A0A1L8DR08 A0A182Q255 A0A1L8DQP1 A0A2P8ZLY1 A0A182IPD0 B4J8F0 A0A1Q3G451 A0A1L8DQU5 A0A182MV14 A0A182VNL1 T1PFY5 A0A1L8DQP5 A0A1Q3G433 A0A182Y8D8 B4GAB8 A0A182L619 Q7Q1V0 A0A2S2QTQ5 A0A224XQS0 A0A182ICP7 A0A084VNI9 A0A2M4BML2 T1HM73 A0A2M4AC36 A0A2M3Z177 A0A2M4ACK0 A0A2M4DMQ3 A0A2M4DMN4 A0A1A9X4X6 A0A2M4DMS1 A0A1B6M6K3 A0A1A9XWH4 A0A1J1IRP1 B3MPM5 A0A151WST7 A0A195BWB5 A0A195CCG6 A0A026WYH3 J9K999 B4M8H2 A0A158NE94 A0A1W4UUS4 B4KRT1 B4NZ59 Q9VL27 J3JUA1 A0A336M9C7 Q8MYY3 A0A195FQA7 B3N935 J9JRF8 B4JP37 B4HWH1 B4Q8Q4 A0A1J1HJ44 F4WJD6 A0A1A9VU43 A0A1B0FRD5 A0A2S2ND41 A0A1A9UT81 A0A1A9ZVN6 T1HJA0 A0A151J0R5 A0A1Y1M843 A0A0L7QQI1 U4UPP5 B4G7B6 A0A1A9WLA3
EC Number
3.2.1.-
Pubmed
22118469
26354079
25315136
10731132
12537568
12537572
+ More
12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 17550304 24495485 25348373 26108605 25830018 18057021 15632085 23185243 18362917 19820115 24845553 17510324 29403074 25244985 20966253 12364791 24438588 24508170 21347285 22516182 21719571 28004739 23537049
12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 17550304 24495485 25348373 26108605 25830018 18057021 15632085 23185243 18362917 19820115 24845553 17510324 29403074 25244985 20966253 12364791 24438588 24508170 21347285 22516182 21719571 28004739 23537049
EMBL
DQ311273
ABD36218.1
ODYU01003025
SOQ41275.1
NWSH01001127
PCG72472.1
+ More
AGBW02009513 OWR50638.1 KQ459185 KPJ03256.1 PCG72470.1 PCG72471.1 AE013599 AY069427 AAF58008.2 AAL39572.1 CH480816 EDW48133.1 CH954179 EDV55764.1 CM000362 CM002911 EDX07348.1 KMY94252.1 CM000158 EDW91932.1 GAMC01009600 JAB96955.1 GAKP01016413 JAC42539.1 JRES01000930 KNC27200.1 GDHF01006436 GDHF01002966 JAI45878.1 JAI49348.1 GBXI01008395 JAD05897.1 CH902619 EDV37725.1 KPU77002.1 CM000071 EAL25033.2 KRT01803.1 KQ971338 EFA01818.1 CH963849 EDW73990.1 GBRD01003152 GBRD01003151 JAG62669.1 CH940667 EDW57218.1 KRF77681.1 OUUW01000001 SPP73435.1 GECZ01016138 GECZ01015544 JAS53631.1 JAS54225.1 KK852444 KDR23773.1 NEVH01017454 PNF24276.1 CH477342 EAT43068.1 GFDF01005314 JAV08770.1 AXCN02000647 GFDF01005311 JAV08773.1 PYGN01000019 PSN57511.1 CH916367 EDW02309.1 GFDL01000468 JAV34577.1 GFDF01005313 JAV08771.1 AXCM01001912 KA647732 AFP62361.1 GFDF01005312 JAV08772.1 GFDL01000485 JAV34560.1 CH479181 EDW31870.1 AAAB01008980 EAA14548.3 GGMS01011299 GGMS01014174 MBY80502.1 MBY83377.1 GFTR01006107 JAW10319.1 APCN01005736 ATLV01014759 KE524984 KFB39533.1 GGFJ01005176 MBW54317.1 ACPB03002860 GGFK01004999 MBW38320.1 GGFM01001531 MBW22282.1 GGFK01005007 MBW38328.1 GGFL01014654 MBW78832.1 GGFL01014652 MBW78830.1 GGFL01014653 MBW78831.1 GEBQ01008421 JAT31556.1 CVRI01000058 CRL02887.1 CH902620 EDV32273.1 KQ982769 KYQ50857.1 KQ976401 KYM92575.1 KQ978023 KYM97921.1 KK107079 EZA60164.1 ABLF02030053 CH940654 EDW57498.2 ADTU01013175 CH933808 EDW08351.1 CM000157 EDW88754.1 AE014134 BT044084 AAF52871.2 ACH92149.1 BT126814 AEE61776.1 UFQS01000272 UFQT01000272 SSX02272.1 SSX22648.1 AY113489 AAM29494.1 KQ981335 KYN42658.1 CH954177 EDV58470.2 ABLF02027051 ABLF02027053 CH916372 EDV99462.1 CH480818 EDW52366.1 CM000361 CM002910 EDX04469.1 KMY89418.1 CVRI01000001 CRK86297.1 GL888182 EGI65714.1 CCAG010015025 GGMR01002419 MBY15038.1 ACPB03006489 KQ980636 KYN14937.1 GEZM01037887 JAV81864.1 KQ414786 KOC60877.1 KB632308 ERL91995.1 CH479180 EDW28370.1
AGBW02009513 OWR50638.1 KQ459185 KPJ03256.1 PCG72470.1 PCG72471.1 AE013599 AY069427 AAF58008.2 AAL39572.1 CH480816 EDW48133.1 CH954179 EDV55764.1 CM000362 CM002911 EDX07348.1 KMY94252.1 CM000158 EDW91932.1 GAMC01009600 JAB96955.1 GAKP01016413 JAC42539.1 JRES01000930 KNC27200.1 GDHF01006436 GDHF01002966 JAI45878.1 JAI49348.1 GBXI01008395 JAD05897.1 CH902619 EDV37725.1 KPU77002.1 CM000071 EAL25033.2 KRT01803.1 KQ971338 EFA01818.1 CH963849 EDW73990.1 GBRD01003152 GBRD01003151 JAG62669.1 CH940667 EDW57218.1 KRF77681.1 OUUW01000001 SPP73435.1 GECZ01016138 GECZ01015544 JAS53631.1 JAS54225.1 KK852444 KDR23773.1 NEVH01017454 PNF24276.1 CH477342 EAT43068.1 GFDF01005314 JAV08770.1 AXCN02000647 GFDF01005311 JAV08773.1 PYGN01000019 PSN57511.1 CH916367 EDW02309.1 GFDL01000468 JAV34577.1 GFDF01005313 JAV08771.1 AXCM01001912 KA647732 AFP62361.1 GFDF01005312 JAV08772.1 GFDL01000485 JAV34560.1 CH479181 EDW31870.1 AAAB01008980 EAA14548.3 GGMS01011299 GGMS01014174 MBY80502.1 MBY83377.1 GFTR01006107 JAW10319.1 APCN01005736 ATLV01014759 KE524984 KFB39533.1 GGFJ01005176 MBW54317.1 ACPB03002860 GGFK01004999 MBW38320.1 GGFM01001531 MBW22282.1 GGFK01005007 MBW38328.1 GGFL01014654 MBW78832.1 GGFL01014652 MBW78830.1 GGFL01014653 MBW78831.1 GEBQ01008421 JAT31556.1 CVRI01000058 CRL02887.1 CH902620 EDV32273.1 KQ982769 KYQ50857.1 KQ976401 KYM92575.1 KQ978023 KYM97921.1 KK107079 EZA60164.1 ABLF02030053 CH940654 EDW57498.2 ADTU01013175 CH933808 EDW08351.1 CM000157 EDW88754.1 AE014134 BT044084 AAF52871.2 ACH92149.1 BT126814 AEE61776.1 UFQS01000272 UFQT01000272 SSX02272.1 SSX22648.1 AY113489 AAM29494.1 KQ981335 KYN42658.1 CH954177 EDV58470.2 ABLF02027051 ABLF02027053 CH916372 EDV99462.1 CH480818 EDW52366.1 CM000361 CM002910 EDX04469.1 KMY89418.1 CVRI01000001 CRK86297.1 GL888182 EGI65714.1 CCAG010015025 GGMR01002419 MBY15038.1 ACPB03006489 KQ980636 KYN14937.1 GEZM01037887 JAV81864.1 KQ414786 KOC60877.1 KB632308 ERL91995.1 CH479180 EDW28370.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000095301
UP000000803
UP000001292
+ More
UP000008711 UP000000304 UP000002282 UP000095300 UP000192221 UP000037069 UP000007801 UP000001819 UP000007266 UP000092445 UP000007798 UP000008792 UP000268350 UP000027135 UP000235965 UP000008820 UP000075886 UP000245037 UP000075880 UP000001070 UP000075883 UP000075903 UP000076408 UP000008744 UP000075882 UP000007062 UP000075840 UP000030765 UP000015103 UP000091820 UP000092443 UP000183832 UP000075809 UP000078540 UP000078542 UP000053097 UP000007819 UP000005205 UP000009192 UP000078541 UP000007755 UP000078200 UP000092444 UP000078492 UP000053825 UP000030742
UP000008711 UP000000304 UP000002282 UP000095300 UP000192221 UP000037069 UP000007801 UP000001819 UP000007266 UP000092445 UP000007798 UP000008792 UP000268350 UP000027135 UP000235965 UP000008820 UP000075886 UP000245037 UP000075880 UP000001070 UP000075883 UP000075903 UP000076408 UP000008744 UP000075882 UP000007062 UP000075840 UP000030765 UP000015103 UP000091820 UP000092443 UP000183832 UP000075809 UP000078540 UP000078542 UP000053097 UP000007819 UP000005205 UP000009192 UP000078541 UP000007755 UP000078200 UP000092444 UP000078492 UP000053825 UP000030742
Interpro
Gene 3D
ProteinModelPortal
Q2F5Z9
A0A2H1VKD1
A0A2A4JM48
A0A212FA86
A0A194QIH9
A0A2A4JM49
+ More
A0A2A4JKI6 A0A1I8N189 Q7K127 B4HSW9 B3NPJ7 B4QHX7 B4P682 A0A1I8PUH1 W8B716 A0A1W4UXD8 A0A034VJ62 A0A0L0C4I6 A0A0K8WDV6 A0A0A1X480 B3MFN5 Q291S9 D1ZZP5 A0A1B0AF32 B4MPF1 A0A0K8TAY4 B4MFP7 A0A3B0IZ83 A0A1B6FU31 A0A067RL82 A0A2J7Q6R5 Q17A03 A0A1L8DR08 A0A182Q255 A0A1L8DQP1 A0A2P8ZLY1 A0A182IPD0 B4J8F0 A0A1Q3G451 A0A1L8DQU5 A0A182MV14 A0A182VNL1 T1PFY5 A0A1L8DQP5 A0A1Q3G433 A0A182Y8D8 B4GAB8 A0A182L619 Q7Q1V0 A0A2S2QTQ5 A0A224XQS0 A0A182ICP7 A0A084VNI9 A0A2M4BML2 T1HM73 A0A2M4AC36 A0A2M3Z177 A0A2M4ACK0 A0A2M4DMQ3 A0A2M4DMN4 A0A1A9X4X6 A0A2M4DMS1 A0A1B6M6K3 A0A1A9XWH4 A0A1J1IRP1 B3MPM5 A0A151WST7 A0A195BWB5 A0A195CCG6 A0A026WYH3 J9K999 B4M8H2 A0A158NE94 A0A1W4UUS4 B4KRT1 B4NZ59 Q9VL27 J3JUA1 A0A336M9C7 Q8MYY3 A0A195FQA7 B3N935 J9JRF8 B4JP37 B4HWH1 B4Q8Q4 A0A1J1HJ44 F4WJD6 A0A1A9VU43 A0A1B0FRD5 A0A2S2ND41 A0A1A9UT81 A0A1A9ZVN6 T1HJA0 A0A151J0R5 A0A1Y1M843 A0A0L7QQI1 U4UPP5 B4G7B6 A0A1A9WLA3
A0A2A4JKI6 A0A1I8N189 Q7K127 B4HSW9 B3NPJ7 B4QHX7 B4P682 A0A1I8PUH1 W8B716 A0A1W4UXD8 A0A034VJ62 A0A0L0C4I6 A0A0K8WDV6 A0A0A1X480 B3MFN5 Q291S9 D1ZZP5 A0A1B0AF32 B4MPF1 A0A0K8TAY4 B4MFP7 A0A3B0IZ83 A0A1B6FU31 A0A067RL82 A0A2J7Q6R5 Q17A03 A0A1L8DR08 A0A182Q255 A0A1L8DQP1 A0A2P8ZLY1 A0A182IPD0 B4J8F0 A0A1Q3G451 A0A1L8DQU5 A0A182MV14 A0A182VNL1 T1PFY5 A0A1L8DQP5 A0A1Q3G433 A0A182Y8D8 B4GAB8 A0A182L619 Q7Q1V0 A0A2S2QTQ5 A0A224XQS0 A0A182ICP7 A0A084VNI9 A0A2M4BML2 T1HM73 A0A2M4AC36 A0A2M3Z177 A0A2M4ACK0 A0A2M4DMQ3 A0A2M4DMN4 A0A1A9X4X6 A0A2M4DMS1 A0A1B6M6K3 A0A1A9XWH4 A0A1J1IRP1 B3MPM5 A0A151WST7 A0A195BWB5 A0A195CCG6 A0A026WYH3 J9K999 B4M8H2 A0A158NE94 A0A1W4UUS4 B4KRT1 B4NZ59 Q9VL27 J3JUA1 A0A336M9C7 Q8MYY3 A0A195FQA7 B3N935 J9JRF8 B4JP37 B4HWH1 B4Q8Q4 A0A1J1HJ44 F4WJD6 A0A1A9VU43 A0A1B0FRD5 A0A2S2ND41 A0A1A9UT81 A0A1A9ZVN6 T1HJA0 A0A151J0R5 A0A1Y1M843 A0A0L7QQI1 U4UPP5 B4G7B6 A0A1A9WLA3
PDB
1KTC
E-value=6.06757e-116,
Score=1068
Ontologies
PATHWAY
00052
Galactose metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00600 Sphingolipid metabolism - Bombyx mori (domestic silkworm)
00603 Glycosphingolipid biosynthesis - globo and isoglobo series - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00600 Sphingolipid metabolism - Bombyx mori (domestic silkworm)
00603 Glycosphingolipid biosynthesis - globo and isoglobo series - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
GO
PANTHER
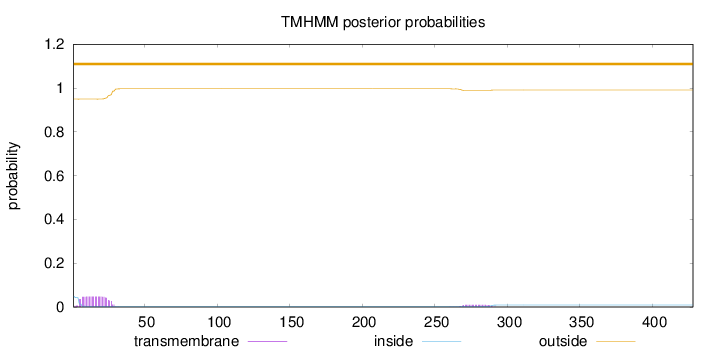
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.989151
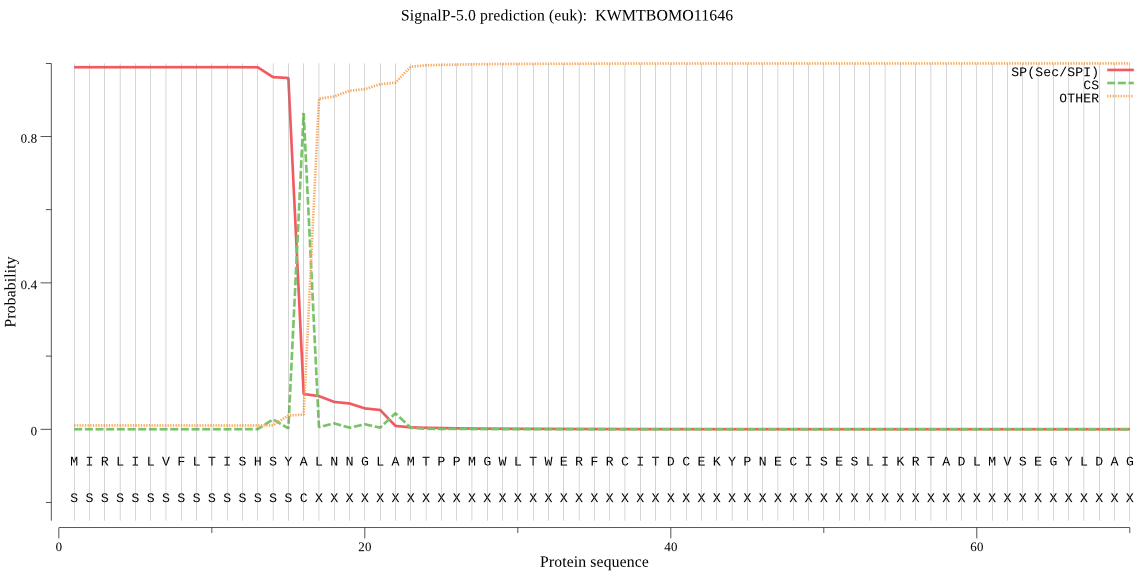
Length:
428
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.24901
Exp number, first 60 AAs:
1.04202
Total prob of N-in:
0.04998
outside
1 - 428
Population Genetic Test Statistics
Pi
27.92622
Theta
22.541395
Tajima's D
-0.642981
CLR
0.720347
CSRT
0.207389630518474
Interpretation
Uncertain
