Pre Gene Modal
BGIBMGA004113
Annotation
PREDICTED:_protein_PRRC1-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.014
Sequence
CDS
ATGAGTTCTGATAGAAAAAAGATTCCTGACCCTCCGACACCAGTTCCTTCAACTGGCAATTTGCTTTCAAATGTAGCACCACCGCTTCAACTTCCAAATTTTGTGACTGCTGTCGCTGAACCAATGCCTACGTCCCAGCGTGAACCGGTGGAGCATCCAGTCCCAGTTCAGCCAATAGTAATACAACAACCTACGAAGATAGATCAGCAAGCATCAGAGAAGTTTTGCCCTGCAATCCCGCCTAGTAAAGGAACACCATTAATGTCTATACGATCACCCGAAGACTCGGAGACTGCACAAACTCCAGAATCCATACCAGAAATTTCACCCGAGATGGATTCATTCAGTGATATAATGCCAGGCACCGGGTTGCTGACGTGGGTGAAGGGTGCAGTTGCCAGTGGTGGTATTCTACAAAGAGTAGCCGAGAAGGCGAAGAATTCAGTAGATTCAATGATCACTACCCTTGATCCACAGATGAAGGAATATCTTAAAAGCGGAGGAGATCTGTACATAGTGGTCGCATCGGATAAAGACGTGAAAGTATCACCAATAAGAGAAGCTTTTCAGACAGTTTTTGGAAAGGCGACTGTGGTAGGTTTCGCGGCTCAGTCTGACGTGACAGCGGCTCAGCCGGTCGGCTATGCGGCCGCTTATGCAGCAGCGAAACAGAGGTTAGCAGATCTTCGGTCCAGACATTCGGAAATAACAAACAATGTTCCCCTAGTGGCCGTTGAGAGTTTCTTGCACGAAGCGATCGCAGACAGATGGTACGAGATGTCGTTGATGATCCTGTCGCAGCCGTCTCTAGGCGTTGAACTGCAAGTGCAGTCGCAGGGCGCGGGCGTGCCGGGCGCCGCCGTGTCCGCCGCGGCCGCCGCCACCGCGCCCTCCTACCCGCACGCGCAGACCGGCTACAGCCTCACCGTAGGATCCATCATGGCCGACAGCCTACAGGTCCCGCACACCGACTGGCAAGAGGCGTCGACCGGCGTGTCTAGGAGGGAAATTCTTTTGTTAGCGGCTAAGTCACTGGCGGGCTCATACAAACGGGTACTAGCGGTATCCGCTGCCGAGACCTAA
Protein
MSSDRKKIPDPPTPVPSTGNLLSNVAPPLQLPNFVTAVAEPMPTSQREPVEHPVPVQPIVIQQPTKIDQQASEKFCPAIPPSKGTPLMSIRSPEDSETAQTPESIPEISPEMDSFSDIMPGTGLLTWVKGAVASGGILQRVAEKAKNSVDSMITTLDPQMKEYLKSGGDLYIVVASDKDVKVSPIREAFQTVFGKATVVGFAAQSDVTAAQPVGYAAAYAAAKQRLADLRSRHSEITNNVPLVAVESFLHEAIADRWYEMSLMILSQPSLGVELQVQSQGAGVPGAAVSAAAAATAPSYPHAQTGYSLTVGSIMADSLQVPHTDWQEASTGVSRREILLLAAKSLAGSYKRVLAVSAAET
Summary
Uniprot
H9J3M6
A0A2A4J2S4
A0A2H1VPD0
A0A194QCK6
A0A212EGY8
S4PDY1
+ More
A0A2J7QJR3 A0A2J7QJR7 A0A2J7QJT0 A0A088A622 B0WFG2 A0A182IW56 A0A0N0BKM1 A0A2M3Z7L9 A0A0N1I8I7 A0A1S4FB56 A0A2M4CXP2 Q179E7 A0A2M4BQ42 A0A2M4BQ45 A0A2M4BQ44 A0A2J7QJS5 A0A182F3J2 A0A1B0GL09 A0A182VDQ2 A0A2M4AFS6 U5EQY6 A0A2A3EKD9 A7UVA8 A0A1S4H0I2 A0A182G1D6 A0A0T6B1P0 A0A1Q3FD28 A0A310SN30 A0A1Y1M0A2 A0A1Q3FD05 A0A1Q3FD45 A0A154P5V3 A0A0L7R200 A0A1W4XE74 A0A1L8DBW6 K7J0G7 A0A1L8DBT7 A0A182PE77 A0A182MEN7 A0A182Q5X5 W5JAQ9 A0A1L8DC65 A0A182WW27 A0A182HGI4 A0A182L6C6 A0A182VUF7 A0A182TN43 A0A0P5ZEX9 A0A1B6DQF6 A0A182SKP2 A0A0P6EYF6 A0A0P5UE99 A0A182YRE8 A0A182MYX4 A0A182RIQ4 A0A182KBQ2 A0A2L2Y9F8 A0A084WH22 A0A0P5VT27 K1Q157 A0A1B6F6D3 A0A2P8YDG9 A0A3B3SY45 A0A3B3SYT6 A0A1B6C033 A0A2D0SRJ3 A0A1B6MA66 A0A1S3M8V0 W5L0Z0 A0A3B1IQK0
A0A2J7QJR3 A0A2J7QJR7 A0A2J7QJT0 A0A088A622 B0WFG2 A0A182IW56 A0A0N0BKM1 A0A2M3Z7L9 A0A0N1I8I7 A0A1S4FB56 A0A2M4CXP2 Q179E7 A0A2M4BQ42 A0A2M4BQ45 A0A2M4BQ44 A0A2J7QJS5 A0A182F3J2 A0A1B0GL09 A0A182VDQ2 A0A2M4AFS6 U5EQY6 A0A2A3EKD9 A7UVA8 A0A1S4H0I2 A0A182G1D6 A0A0T6B1P0 A0A1Q3FD28 A0A310SN30 A0A1Y1M0A2 A0A1Q3FD05 A0A1Q3FD45 A0A154P5V3 A0A0L7R200 A0A1W4XE74 A0A1L8DBW6 K7J0G7 A0A1L8DBT7 A0A182PE77 A0A182MEN7 A0A182Q5X5 W5JAQ9 A0A1L8DC65 A0A182WW27 A0A182HGI4 A0A182L6C6 A0A182VUF7 A0A182TN43 A0A0P5ZEX9 A0A1B6DQF6 A0A182SKP2 A0A0P6EYF6 A0A0P5UE99 A0A182YRE8 A0A182MYX4 A0A182RIQ4 A0A182KBQ2 A0A2L2Y9F8 A0A084WH22 A0A0P5VT27 K1Q157 A0A1B6F6D3 A0A2P8YDG9 A0A3B3SY45 A0A3B3SYT6 A0A1B6C033 A0A2D0SRJ3 A0A1B6MA66 A0A1S3M8V0 W5L0Z0 A0A3B1IQK0
Pubmed
EMBL
BABH01021636
NWSH01003621
PCG66036.1
ODYU01003470
SOQ42302.1
KQ459185
+ More
KPJ03273.1 AGBW02014995 OWR40753.1 GAIX01003446 JAA89114.1 NEVH01013549 PNF28833.1 PNF28830.1 PNF28829.1 DS231917 EDS26199.1 KQ435697 KOX80792.1 GGFM01003775 MBW24526.1 KQ460772 KPJ12493.1 GGFL01005916 MBW70094.1 CH477353 EAT42859.1 GGFJ01006065 MBW55206.1 GGFJ01006066 MBW55207.1 GGFJ01006064 MBW55205.1 PNF28832.1 AJWK01034038 AJWK01034039 GGFK01006324 MBW39645.1 GANO01004029 JAB55842.1 KZ288229 PBC31656.1 AAAB01008984 EDO63407.1 JXUM01136543 KQ568445 KXJ68999.1 LJIG01016272 KRT81099.1 GFDL01009578 JAV25467.1 KQ759828 OAD62655.1 GEZM01044892 GEZM01044891 GEZM01044890 GEZM01044889 JAV78060.1 GFDL01009564 JAV25481.1 GFDL01009559 JAV25486.1 KQ434809 KZC06578.1 KQ414667 KOC64859.1 GFDF01010126 JAV03958.1 GFDF01010300 JAV03784.1 AXCM01001281 AXCN02000815 ADMH02001788 ETN61086.1 GFDF01010127 JAV03957.1 APCN01004236 GDIP01044612 JAM59103.1 GEDC01009400 JAS27898.1 GDIQ01055769 JAN38968.1 GDIP01115656 JAL88058.1 IAAA01012243 IAAA01012244 LAA03980.1 ATLV01023778 KE525346 KFB49516.1 GDIP01095857 JAM07858.1 JH816331 EKC22530.1 GECZ01023991 JAS45778.1 PYGN01000681 PSN42301.1 GEDC01030574 JAS06724.1 GEBQ01007161 GEBQ01002004 JAT32816.1 JAT37973.1
KPJ03273.1 AGBW02014995 OWR40753.1 GAIX01003446 JAA89114.1 NEVH01013549 PNF28833.1 PNF28830.1 PNF28829.1 DS231917 EDS26199.1 KQ435697 KOX80792.1 GGFM01003775 MBW24526.1 KQ460772 KPJ12493.1 GGFL01005916 MBW70094.1 CH477353 EAT42859.1 GGFJ01006065 MBW55206.1 GGFJ01006066 MBW55207.1 GGFJ01006064 MBW55205.1 PNF28832.1 AJWK01034038 AJWK01034039 GGFK01006324 MBW39645.1 GANO01004029 JAB55842.1 KZ288229 PBC31656.1 AAAB01008984 EDO63407.1 JXUM01136543 KQ568445 KXJ68999.1 LJIG01016272 KRT81099.1 GFDL01009578 JAV25467.1 KQ759828 OAD62655.1 GEZM01044892 GEZM01044891 GEZM01044890 GEZM01044889 JAV78060.1 GFDL01009564 JAV25481.1 GFDL01009559 JAV25486.1 KQ434809 KZC06578.1 KQ414667 KOC64859.1 GFDF01010126 JAV03958.1 GFDF01010300 JAV03784.1 AXCM01001281 AXCN02000815 ADMH02001788 ETN61086.1 GFDF01010127 JAV03957.1 APCN01004236 GDIP01044612 JAM59103.1 GEDC01009400 JAS27898.1 GDIQ01055769 JAN38968.1 GDIP01115656 JAL88058.1 IAAA01012243 IAAA01012244 LAA03980.1 ATLV01023778 KE525346 KFB49516.1 GDIP01095857 JAM07858.1 JH816331 EKC22530.1 GECZ01023991 JAS45778.1 PYGN01000681 PSN42301.1 GEDC01030574 JAS06724.1 GEBQ01007161 GEBQ01002004 JAT32816.1 JAT37973.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000235965
UP000005203
+ More
UP000002320 UP000075880 UP000053105 UP000053240 UP000008820 UP000069272 UP000092461 UP000075903 UP000242457 UP000007062 UP000069940 UP000249989 UP000076502 UP000053825 UP000192223 UP000002358 UP000075885 UP000075883 UP000075886 UP000000673 UP000076407 UP000075840 UP000075882 UP000075920 UP000075902 UP000075901 UP000076408 UP000075884 UP000075900 UP000075881 UP000030765 UP000005408 UP000245037 UP000261540 UP000221080 UP000087266 UP000018467
UP000002320 UP000075880 UP000053105 UP000053240 UP000008820 UP000069272 UP000092461 UP000075903 UP000242457 UP000007062 UP000069940 UP000249989 UP000076502 UP000053825 UP000192223 UP000002358 UP000075885 UP000075883 UP000075886 UP000000673 UP000076407 UP000075840 UP000075882 UP000075920 UP000075902 UP000075901 UP000076408 UP000075884 UP000075900 UP000075881 UP000030765 UP000005408 UP000245037 UP000261540 UP000221080 UP000087266 UP000018467
PRIDE
Pfam
PF01931 NTPase_I-T
SUPFAM
SSF52972
SSF52972
Gene 3D
ProteinModelPortal
H9J3M6
A0A2A4J2S4
A0A2H1VPD0
A0A194QCK6
A0A212EGY8
S4PDY1
+ More
A0A2J7QJR3 A0A2J7QJR7 A0A2J7QJT0 A0A088A622 B0WFG2 A0A182IW56 A0A0N0BKM1 A0A2M3Z7L9 A0A0N1I8I7 A0A1S4FB56 A0A2M4CXP2 Q179E7 A0A2M4BQ42 A0A2M4BQ45 A0A2M4BQ44 A0A2J7QJS5 A0A182F3J2 A0A1B0GL09 A0A182VDQ2 A0A2M4AFS6 U5EQY6 A0A2A3EKD9 A7UVA8 A0A1S4H0I2 A0A182G1D6 A0A0T6B1P0 A0A1Q3FD28 A0A310SN30 A0A1Y1M0A2 A0A1Q3FD05 A0A1Q3FD45 A0A154P5V3 A0A0L7R200 A0A1W4XE74 A0A1L8DBW6 K7J0G7 A0A1L8DBT7 A0A182PE77 A0A182MEN7 A0A182Q5X5 W5JAQ9 A0A1L8DC65 A0A182WW27 A0A182HGI4 A0A182L6C6 A0A182VUF7 A0A182TN43 A0A0P5ZEX9 A0A1B6DQF6 A0A182SKP2 A0A0P6EYF6 A0A0P5UE99 A0A182YRE8 A0A182MYX4 A0A182RIQ4 A0A182KBQ2 A0A2L2Y9F8 A0A084WH22 A0A0P5VT27 K1Q157 A0A1B6F6D3 A0A2P8YDG9 A0A3B3SY45 A0A3B3SYT6 A0A1B6C033 A0A2D0SRJ3 A0A1B6MA66 A0A1S3M8V0 W5L0Z0 A0A3B1IQK0
A0A2J7QJR3 A0A2J7QJR7 A0A2J7QJT0 A0A088A622 B0WFG2 A0A182IW56 A0A0N0BKM1 A0A2M3Z7L9 A0A0N1I8I7 A0A1S4FB56 A0A2M4CXP2 Q179E7 A0A2M4BQ42 A0A2M4BQ45 A0A2M4BQ44 A0A2J7QJS5 A0A182F3J2 A0A1B0GL09 A0A182VDQ2 A0A2M4AFS6 U5EQY6 A0A2A3EKD9 A7UVA8 A0A1S4H0I2 A0A182G1D6 A0A0T6B1P0 A0A1Q3FD28 A0A310SN30 A0A1Y1M0A2 A0A1Q3FD05 A0A1Q3FD45 A0A154P5V3 A0A0L7R200 A0A1W4XE74 A0A1L8DBW6 K7J0G7 A0A1L8DBT7 A0A182PE77 A0A182MEN7 A0A182Q5X5 W5JAQ9 A0A1L8DC65 A0A182WW27 A0A182HGI4 A0A182L6C6 A0A182VUF7 A0A182TN43 A0A0P5ZEX9 A0A1B6DQF6 A0A182SKP2 A0A0P6EYF6 A0A0P5UE99 A0A182YRE8 A0A182MYX4 A0A182RIQ4 A0A182KBQ2 A0A2L2Y9F8 A0A084WH22 A0A0P5VT27 K1Q157 A0A1B6F6D3 A0A2P8YDG9 A0A3B3SY45 A0A3B3SYT6 A0A1B6C033 A0A2D0SRJ3 A0A1B6MA66 A0A1S3M8V0 W5L0Z0 A0A3B1IQK0
Ontologies
PANTHER
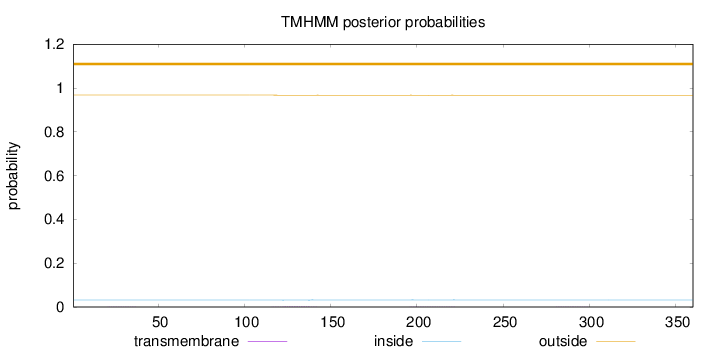
Topology
Length:
360
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0489400000000001
Exp number, first 60 AAs:
0.00279
Total prob of N-in:
0.03204
outside
1 - 360
Population Genetic Test Statistics
Pi
162.617744
Theta
162.369202
Tajima's D
0.081537
CLR
1.354
CSRT
0.392030398480076
Interpretation
Uncertain
