Gene
KWMTBOMO11616 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004010
Annotation
PREDICTED:_sorting_nexin-2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.846 Nuclear Reliability : 1.649
Sequence
CDS
ATGTCGGCGGGCCCAGATACTCCGCCATTTAGCACCGTTGACATCAACAATGATAGCCGAGATGACGAGGATATTTTTGCGTCCGCTGTACAGGAAGTCAGTCTGGATCCAGAAGTGAATGGAGCTCGTGAGGGTCTGGAGAAAGTTTCCATCGATGACACTCCAGCCTCAATAAGTACCTCTCTCAACAGTCCAGTTATGGAAGAGATTGCCACAGAACGTTCAAATAATATCATAATAACGGTAACGGAACCACAAAAAATTGGAGAAGGCATGAGTTCGTACGTCGCGTATCGAGTATTCACCAAAACAAACATGCCGATATTTAGTAAGCACGAATTCGCTGTGCTCAGACGTTTCAGTGACTTCTTGGGCTTACACGAAAAGCTCACTGAGAAATATCTGCGCTCCGGAAGGATAATACCACCTGCACCTGAAAAAAGCATAGTTGGCACAACGAAACTAAAGATGACATCGACCCCTTCAACGGAGAACGTGAATGGAAGCGTATCGAGCGCGGGGGTGCAGTCTCAGTTCGTGGAACGTCGGCGGGCCGCTCTAGAGAGGTTCCTTAATCGTGTAGCCCAGCACCCGGTCTTGAGCATCGACCCTGATTTTAGAGAATTCCTCGAGTCAGGAATGGACCTACCGAAAGCCACAAGTACATCGGCGTTGAGTGGAGCGGGAATGTTACGTCTCTTCAATAAAGTCGGTGAGACCGTCAACAAGATCACTTATAGAATGGACGAGGCTGATCCGTGGTTCGAGGAGCGCGCGCTTCGCGTGGAGACGCTGGAGGCGTGCCTGCGGCGGCTCGGCGGCGCGTGCGAGGCGCTGGCGGGCGAGCGACGCGAGCTGGCCGTGCGCACGCACGACGCGGCGCGCGCGCACGCCTCACTGTCCGGCGCCGACCCGCACGCGCCCCTCTCCCGCGCGCTCTCCCACGTCGCCGACCTGCACGAGAAGATTGAAAACTTGAGACTGGAACAATCAAACACGGACTTCTTCATTCTGTCTGAACACATCAAAGATTACCTCGGGCTTATCGGATCAATCAAAGACGTATTCCATGAACGAGTAAAGGTGTTCCAAAACTGGCAACACGCTCAAATGCAGCTCACTAAAAGGCGCGAGAACAAAGCCAAGGCTGAACTCTCGAACCGACCGGATAAAGCTGAACAAGCAGCCAATGAAATAGTTGAGTGGGAAACAAAAGTCGAACGCAATCAACAAGAGTTCGACTCGATATCGCGCGTCATCAAGAAGGAAATCGAGCGATGGGAGGAGATCCGGGTCGATGAGTTGCGGGCGACGCTGCTGCACTACCTCGAGGAACACATGAAGCATCAAGCTCAGGCGATTAGATATTGGGACGCGTTCCTTCCCGAAGCGCGCGCCATCAAATGA
Protein
MSAGPDTPPFSTVDINNDSRDDEDIFASAVQEVSLDPEVNGAREGLEKVSIDDTPASISTSLNSPVMEEIATERSNNIIITVTEPQKIGEGMSSYVAYRVFTKTNMPIFSKHEFAVLRRFSDFLGLHEKLTEKYLRSGRIIPPAPEKSIVGTTKLKMTSTPSTENVNGSVSSAGVQSQFVERRRAALERFLNRVAQHPVLSIDPDFREFLESGMDLPKATSTSALSGAGMLRLFNKVGETVNKITYRMDEADPWFEERALRVETLEACLRRLGGACEALAGERRELAVRTHDAARAHASLSGADPHAPLSRALSHVADLHEKIENLRLEQSNTDFFILSEHIKDYLGLIGSIKDVFHERVKVFQNWQHAQMQLTKRRENKAKAELSNRPDKAEQAANEIVEWETKVERNQQEFDSISRVIKKEIERWEEIRVDELRATLLHYLEEHMKHQAQAIRYWDAFLPEARAIK
Summary
Uniprot
H9J3C3
A0A3S2LUY4
A0A194QCX7
A0A2H1VNB2
A0A0N0PBW7
A0A212EH05
+ More
A0A067RBF7 D6WRQ8 V5GSX9 A0A2J7QJQ0 A0A1W4XJJ1 A0A1Y1LWZ1 A0A195D304 A0A2A3EMS4 A0A195E4B1 A0A195FJ95 A0A182WW28 A0A0J7L9J4 A0A0M8ZUA4 A0A182V2Y7 A0A182L6C4 A0A151X240 Q7PVK3 A0A154P3Z7 E9IV11 E2AJW8 A0A182TZW5 E2B387 A0A0K8TMI1 A0A182PE78 N6T3D5 A0A2P8YDG7 F4W6H6 A0A182KBQ1 A0A182QGB9 A0A084WH21 A0A195AVU3 A0A158P283 A0A1Q3G1D2 A0A182JKB1 A0A182MYX3 A0A2M4BMP9 A0A2M3Z559 A0A2M4BNK6 U5ERM9 W5J9F1 A0A1Q3G1D0 A0A232EVS2 A0A1Q3G1B4 A0A2M4AKU6 A0A2M4AKQ0 A0A182F3J3 A0A088AL66 A0A1L8DV84 A0A1S4FB77 A0A0C9QF80 A0A026WWY6 Q179F0 A0A182MW50 A0A1B6LMV1 A0A182T568 A0A182VUF8 B4KJL4 B4LSG2 A0A0T6B186 A0A182HGI3 B4JBU4 A0A224XMN1 A0A182RIQ3 A0A182YRE9 T1ID17 B3MVG7 B3N320 Q179E9 B4G861 A0A0C9QU20 A0A034WRH0 A0A1I8MLZ7 B4N106 Q9VQQ6 A0A0P4W091 A0A0J9TEQ4 A0A1I8PE38 A0A1W4V724 B4NXW5 A0A0K8U4J8 A0A0A1WJQ5 A0A2H8TDV7 Q8MS29 B4INK0 A0A1I8PDZ4 Q29N71 A0A2S2RAL3 A0A0M4E0X2 A0A1J1IRC0 W8BJF6 A0A1D2N4U2 J9JMW6 A0A1A9WKK7 A0A0L0CMJ6
A0A067RBF7 D6WRQ8 V5GSX9 A0A2J7QJQ0 A0A1W4XJJ1 A0A1Y1LWZ1 A0A195D304 A0A2A3EMS4 A0A195E4B1 A0A195FJ95 A0A182WW28 A0A0J7L9J4 A0A0M8ZUA4 A0A182V2Y7 A0A182L6C4 A0A151X240 Q7PVK3 A0A154P3Z7 E9IV11 E2AJW8 A0A182TZW5 E2B387 A0A0K8TMI1 A0A182PE78 N6T3D5 A0A2P8YDG7 F4W6H6 A0A182KBQ1 A0A182QGB9 A0A084WH21 A0A195AVU3 A0A158P283 A0A1Q3G1D2 A0A182JKB1 A0A182MYX3 A0A2M4BMP9 A0A2M3Z559 A0A2M4BNK6 U5ERM9 W5J9F1 A0A1Q3G1D0 A0A232EVS2 A0A1Q3G1B4 A0A2M4AKU6 A0A2M4AKQ0 A0A182F3J3 A0A088AL66 A0A1L8DV84 A0A1S4FB77 A0A0C9QF80 A0A026WWY6 Q179F0 A0A182MW50 A0A1B6LMV1 A0A182T568 A0A182VUF8 B4KJL4 B4LSG2 A0A0T6B186 A0A182HGI3 B4JBU4 A0A224XMN1 A0A182RIQ3 A0A182YRE9 T1ID17 B3MVG7 B3N320 Q179E9 B4G861 A0A0C9QU20 A0A034WRH0 A0A1I8MLZ7 B4N106 Q9VQQ6 A0A0P4W091 A0A0J9TEQ4 A0A1I8PE38 A0A1W4V724 B4NXW5 A0A0K8U4J8 A0A0A1WJQ5 A0A2H8TDV7 Q8MS29 B4INK0 A0A1I8PDZ4 Q29N71 A0A2S2RAL3 A0A0M4E0X2 A0A1J1IRC0 W8BJF6 A0A1D2N4U2 J9JMW6 A0A1A9WKK7 A0A0L0CMJ6
Pubmed
19121390
26354079
22118469
24845553
18362917
19820115
+ More
28004739 20966253 12364791 14747013 17210077 21282665 20798317 26369729 23537049 29403074 21719571 24438588 21347285 20920257 23761445 28648823 24508170 30249741 17510324 17994087 25244985 25348373 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 22936249 17550304 25830018 15632085 24495485 27289101 26108605
28004739 20966253 12364791 14747013 17210077 21282665 20798317 26369729 23537049 29403074 21719571 24438588 21347285 20920257 23761445 28648823 24508170 30249741 17510324 17994087 25244985 25348373 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 22936249 17550304 25830018 15632085 24495485 27289101 26108605
EMBL
BABH01021635
BABH01021636
RSAL01000182
RVE44899.1
KQ459185
KPJ03274.1
+ More
ODYU01003470 SOQ42303.1 KQ460772 KPJ12494.1 AGBW02014995 OWR40754.1 KK852809 KDR15997.1 KQ971354 EFA05966.1 GALX01001177 JAB67289.1 NEVH01013549 PNF28821.1 GEZM01044892 GEZM01044891 GEZM01044890 GEZM01044889 JAV78063.1 KQ976885 KYN07277.1 KZ288209 PBC32990.1 KQ979657 KYN19921.1 KQ981523 KYN40332.1 LBMM01000122 KMR04851.1 KQ435843 KOX71285.1 KQ982580 KYQ54479.1 AAAB01008984 EAA14938.3 KQ434809 KZC06665.1 GL766116 EFZ15557.1 GL440100 EFN66284.1 GL445305 EFN89869.1 GDAI01002026 JAI15577.1 APGK01045628 KB741037 KB632319 ENN74654.1 ERL92300.1 PYGN01000681 PSN42299.1 GL887707 EGI70311.1 AXCN02000815 ATLV01023778 KE525346 KFB49515.1 KQ976731 KYM76182.1 ADTU01007023 GFDL01001445 JAV33600.1 GGFJ01005218 MBW54359.1 GGFM01002906 MBW23657.1 GGFJ01005217 MBW54358.1 GANO01003672 JAB56199.1 ADMH02001788 ETN61087.1 GFDL01001442 JAV33603.1 NNAY01001961 OXU22445.1 GFDL01001450 JAV33595.1 GGFK01008080 MBW41401.1 GGFK01008013 MBW41334.1 GFDF01003748 JAV10336.1 GBYB01000837 GBYB01002049 JAG70604.1 JAG71816.1 KK107077 QOIP01000010 EZA60562.1 RLU17753.1 CH477353 EAT42856.1 AXCM01001281 GEBQ01014906 JAT25071.1 CH933807 EDW11459.1 CH940649 EDW64784.2 LJIG01016272 KRT81098.1 APCN01004236 CH916368 EDW04047.1 GFTR01006584 JAW09842.1 ACPB03002827 CH902624 EDV33232.1 CH954177 EDV57619.1 EAT42857.1 CH479180 EDW28541.1 GBYB01007234 JAG77001.1 GAKP01001748 JAC57204.1 CH963920 EDW78168.1 AE014134 BT133048 AAF51109.1 AEX93134.1 GDKW01000325 JAI56270.1 CM002910 KMY87890.1 CM000157 EDW87536.1 GDHF01030873 JAI21441.1 GBXI01015552 JAC98739.1 GFXV01000482 MBW12287.1 AY119129 AAM50989.1 CH676463 EDW44609.1 CH379060 EAL33472.1 GGMS01017894 MBY87097.1 CP012523 ALC38891.1 CVRI01000055 CRL01630.1 GAMC01005140 GAMC01005138 JAC01416.1 LJIJ01000217 ODN00297.1 ABLF02020714 ABLF02020715 ABLF02046332 JRES01000186 KNC33501.1
ODYU01003470 SOQ42303.1 KQ460772 KPJ12494.1 AGBW02014995 OWR40754.1 KK852809 KDR15997.1 KQ971354 EFA05966.1 GALX01001177 JAB67289.1 NEVH01013549 PNF28821.1 GEZM01044892 GEZM01044891 GEZM01044890 GEZM01044889 JAV78063.1 KQ976885 KYN07277.1 KZ288209 PBC32990.1 KQ979657 KYN19921.1 KQ981523 KYN40332.1 LBMM01000122 KMR04851.1 KQ435843 KOX71285.1 KQ982580 KYQ54479.1 AAAB01008984 EAA14938.3 KQ434809 KZC06665.1 GL766116 EFZ15557.1 GL440100 EFN66284.1 GL445305 EFN89869.1 GDAI01002026 JAI15577.1 APGK01045628 KB741037 KB632319 ENN74654.1 ERL92300.1 PYGN01000681 PSN42299.1 GL887707 EGI70311.1 AXCN02000815 ATLV01023778 KE525346 KFB49515.1 KQ976731 KYM76182.1 ADTU01007023 GFDL01001445 JAV33600.1 GGFJ01005218 MBW54359.1 GGFM01002906 MBW23657.1 GGFJ01005217 MBW54358.1 GANO01003672 JAB56199.1 ADMH02001788 ETN61087.1 GFDL01001442 JAV33603.1 NNAY01001961 OXU22445.1 GFDL01001450 JAV33595.1 GGFK01008080 MBW41401.1 GGFK01008013 MBW41334.1 GFDF01003748 JAV10336.1 GBYB01000837 GBYB01002049 JAG70604.1 JAG71816.1 KK107077 QOIP01000010 EZA60562.1 RLU17753.1 CH477353 EAT42856.1 AXCM01001281 GEBQ01014906 JAT25071.1 CH933807 EDW11459.1 CH940649 EDW64784.2 LJIG01016272 KRT81098.1 APCN01004236 CH916368 EDW04047.1 GFTR01006584 JAW09842.1 ACPB03002827 CH902624 EDV33232.1 CH954177 EDV57619.1 EAT42857.1 CH479180 EDW28541.1 GBYB01007234 JAG77001.1 GAKP01001748 JAC57204.1 CH963920 EDW78168.1 AE014134 BT133048 AAF51109.1 AEX93134.1 GDKW01000325 JAI56270.1 CM002910 KMY87890.1 CM000157 EDW87536.1 GDHF01030873 JAI21441.1 GBXI01015552 JAC98739.1 GFXV01000482 MBW12287.1 AY119129 AAM50989.1 CH676463 EDW44609.1 CH379060 EAL33472.1 GGMS01017894 MBY87097.1 CP012523 ALC38891.1 CVRI01000055 CRL01630.1 GAMC01005140 GAMC01005138 JAC01416.1 LJIJ01000217 ODN00297.1 ABLF02020714 ABLF02020715 ABLF02046332 JRES01000186 KNC33501.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000007266 UP000235965 UP000192223 UP000078542 UP000242457 UP000078492 UP000078541 UP000076407 UP000036403 UP000053105 UP000075903 UP000075882 UP000075809 UP000007062 UP000076502 UP000000311 UP000075902 UP000008237 UP000075885 UP000019118 UP000030742 UP000245037 UP000007755 UP000075881 UP000075886 UP000030765 UP000078540 UP000005205 UP000075880 UP000075884 UP000000673 UP000215335 UP000069272 UP000005203 UP000053097 UP000279307 UP000008820 UP000075883 UP000075901 UP000075920 UP000009192 UP000008792 UP000075840 UP000001070 UP000075900 UP000076408 UP000015103 UP000007801 UP000008711 UP000008744 UP000095301 UP000007798 UP000000803 UP000095300 UP000192221 UP000002282 UP000001292 UP000001819 UP000092553 UP000183832 UP000094527 UP000007819 UP000091820 UP000037069
UP000007266 UP000235965 UP000192223 UP000078542 UP000242457 UP000078492 UP000078541 UP000076407 UP000036403 UP000053105 UP000075903 UP000075882 UP000075809 UP000007062 UP000076502 UP000000311 UP000075902 UP000008237 UP000075885 UP000019118 UP000030742 UP000245037 UP000007755 UP000075881 UP000075886 UP000030765 UP000078540 UP000005205 UP000075880 UP000075884 UP000000673 UP000215335 UP000069272 UP000005203 UP000053097 UP000279307 UP000008820 UP000075883 UP000075901 UP000075920 UP000009192 UP000008792 UP000075840 UP000001070 UP000075900 UP000076408 UP000015103 UP000007801 UP000008711 UP000008744 UP000095301 UP000007798 UP000000803 UP000095300 UP000192221 UP000002282 UP000001292 UP000001819 UP000092553 UP000183832 UP000094527 UP000007819 UP000091820 UP000037069
Interpro
Gene 3D
ProteinModelPortal
H9J3C3
A0A3S2LUY4
A0A194QCX7
A0A2H1VNB2
A0A0N0PBW7
A0A212EH05
+ More
A0A067RBF7 D6WRQ8 V5GSX9 A0A2J7QJQ0 A0A1W4XJJ1 A0A1Y1LWZ1 A0A195D304 A0A2A3EMS4 A0A195E4B1 A0A195FJ95 A0A182WW28 A0A0J7L9J4 A0A0M8ZUA4 A0A182V2Y7 A0A182L6C4 A0A151X240 Q7PVK3 A0A154P3Z7 E9IV11 E2AJW8 A0A182TZW5 E2B387 A0A0K8TMI1 A0A182PE78 N6T3D5 A0A2P8YDG7 F4W6H6 A0A182KBQ1 A0A182QGB9 A0A084WH21 A0A195AVU3 A0A158P283 A0A1Q3G1D2 A0A182JKB1 A0A182MYX3 A0A2M4BMP9 A0A2M3Z559 A0A2M4BNK6 U5ERM9 W5J9F1 A0A1Q3G1D0 A0A232EVS2 A0A1Q3G1B4 A0A2M4AKU6 A0A2M4AKQ0 A0A182F3J3 A0A088AL66 A0A1L8DV84 A0A1S4FB77 A0A0C9QF80 A0A026WWY6 Q179F0 A0A182MW50 A0A1B6LMV1 A0A182T568 A0A182VUF8 B4KJL4 B4LSG2 A0A0T6B186 A0A182HGI3 B4JBU4 A0A224XMN1 A0A182RIQ3 A0A182YRE9 T1ID17 B3MVG7 B3N320 Q179E9 B4G861 A0A0C9QU20 A0A034WRH0 A0A1I8MLZ7 B4N106 Q9VQQ6 A0A0P4W091 A0A0J9TEQ4 A0A1I8PE38 A0A1W4V724 B4NXW5 A0A0K8U4J8 A0A0A1WJQ5 A0A2H8TDV7 Q8MS29 B4INK0 A0A1I8PDZ4 Q29N71 A0A2S2RAL3 A0A0M4E0X2 A0A1J1IRC0 W8BJF6 A0A1D2N4U2 J9JMW6 A0A1A9WKK7 A0A0L0CMJ6
A0A067RBF7 D6WRQ8 V5GSX9 A0A2J7QJQ0 A0A1W4XJJ1 A0A1Y1LWZ1 A0A195D304 A0A2A3EMS4 A0A195E4B1 A0A195FJ95 A0A182WW28 A0A0J7L9J4 A0A0M8ZUA4 A0A182V2Y7 A0A182L6C4 A0A151X240 Q7PVK3 A0A154P3Z7 E9IV11 E2AJW8 A0A182TZW5 E2B387 A0A0K8TMI1 A0A182PE78 N6T3D5 A0A2P8YDG7 F4W6H6 A0A182KBQ1 A0A182QGB9 A0A084WH21 A0A195AVU3 A0A158P283 A0A1Q3G1D2 A0A182JKB1 A0A182MYX3 A0A2M4BMP9 A0A2M3Z559 A0A2M4BNK6 U5ERM9 W5J9F1 A0A1Q3G1D0 A0A232EVS2 A0A1Q3G1B4 A0A2M4AKU6 A0A2M4AKQ0 A0A182F3J3 A0A088AL66 A0A1L8DV84 A0A1S4FB77 A0A0C9QF80 A0A026WWY6 Q179F0 A0A182MW50 A0A1B6LMV1 A0A182T568 A0A182VUF8 B4KJL4 B4LSG2 A0A0T6B186 A0A182HGI3 B4JBU4 A0A224XMN1 A0A182RIQ3 A0A182YRE9 T1ID17 B3MVG7 B3N320 Q179E9 B4G861 A0A0C9QU20 A0A034WRH0 A0A1I8MLZ7 B4N106 Q9VQQ6 A0A0P4W091 A0A0J9TEQ4 A0A1I8PE38 A0A1W4V724 B4NXW5 A0A0K8U4J8 A0A0A1WJQ5 A0A2H8TDV7 Q8MS29 B4INK0 A0A1I8PDZ4 Q29N71 A0A2S2RAL3 A0A0M4E0X2 A0A1J1IRC0 W8BJF6 A0A1D2N4U2 J9JMW6 A0A1A9WKK7 A0A0L0CMJ6
PDB
4FZS
E-value=1.44044e-41,
Score=427
Ontologies
GO
PANTHER
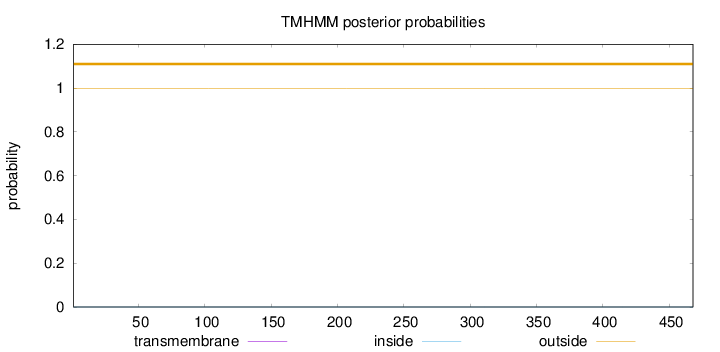
Topology
Length:
468
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00058
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00055
outside
1 - 468
Population Genetic Test Statistics
Pi
18.691413
Theta
21.81529
Tajima's D
-0.792974
CLR
1.176653
CSRT
0.176291185440728
Interpretation
Uncertain
