Pre Gene Modal
BGIBMGA004112
Annotation
PREDICTED:_39S_ribosomal_protein_L13?_mitochondrial_[Papilio_machaon]
Full name
39S ribosomal protein L13, mitochondrial
Location in the cell
Mitochondrial Reliability : 3.052
Sequence
CDS
ATGTCAGCAGCGAAACGAGTACAGCAATGGGCAACATTTGCTCGTAGTTGGCACATATTCGACTGCAAATGGCAAGATCCTTATGAATCCGCTCACGTTATAAAAAAATATTTGATGGGCATGCACAAACCCATATACCATCCTATGAACGATTGTGGAGATGTTGTTGTCTGTATAAATAGTAGAGAGATAGCTCTTCGAGGCGACGAGTGGAGGAAACGTGCTTATTTCCACCACACTGGATACCCTGGAGGTGCTTCGTGGACTTTAGCATGGGAGTTACATAACAAGGATCCAACCATGATCATAAAAAAAGCAGTGTACAGAGCAATGACTGGAAACCTTCAGAGGCGTCATACAATGCAGCGTCTTTTTATATACCCTGGAGAGAATGTACCGGAAGACGTTTTGCAAAATGTTACTAATCAAATTAGACAAATACGTTACGTACCGAAAAGACTAGACCACATACCCGAAGAAGAAGTGAAAAATTATCCCAAAGTCATGGAATACCCAGAAGATTTTGTTGTTAAATAA
Protein
MSAAKRVQQWATFARSWHIFDCKWQDPYESAHVIKKYLMGMHKPIYHPMNDCGDVVVCINSREIALRGDEWRKRAYFHHTGYPGGASWTLAWELHNKDPTMIIKKAVYRAMTGNLQRRHTMQRLFIYPGENVPEDVLQNVTNQIRQIRYVPKRLDHIPEEEVKNYPKVMEYPEDFVVK
Summary
Subunit
Component of the mitochondrial ribosome large subunit (39S) which comprises a 16S rRNA and about 50 distinct proteins (By similarity).
Similarity
Belongs to the universal ribosomal protein uL13 family.
Keywords
Complete proteome
Mitochondrion
Reference proteome
Ribonucleoprotein
Ribosomal protein
Feature
chain 39S ribosomal protein L13, mitochondrial
Uniprot
H9J3M5
A0A2A4J110
A0A0N0PBZ0
A0A194QIJ7
A0A2H1VFC0
A0A182QKX5
+ More
A0A0P8Y5U0 A0A0Q9W9H4 A0A182PTD8 A0A182RR81 A0A1S4H2D3 A0A182V6J5 A0A182XJK2 A0A182KTK3 A0A182HRI0 A0A2M4C174 A0A182YMS5 A0A182JTT1 A0A2M4C136 W5JSH0 A0A2M3Z7N2 A0A2M4AVX9 A0A0R3NSN0 B4G8J0 A0A1Q3FE64 A0A182U7I3 A0A1A9VJC2 A0A1W4V9D9 A0A182NAS6 B0WVS2 A0A182F3W1 A0A1L8DRT2 A0A182W906 B4JB54 D3TRV4 B4I5M1 B4Q8Z6 A0A139WF38 A0A0R1DPC5 A0A0Q5VUJ7 A0A182M460 A0A023EHV2 Q9VJ38 A0A0Q9X221 A0A0Q9X3E8 A0A0K8W6L9 J3JY52 W8C7W6 A0A0A1X9F2 Q17ET7 A0A1A9Y5N8 A0A240PLE7 Q7Q191 A0A1W4WU47 E2AXI1 A0A0L0BZT8 A0A1L8ED45 A0A1Y1MJK0 A0A1L8EDC2 A0A1B0DH47 A0A067RAN4 A0A0M5J9G6 A0A1I8MKJ8 A0A1I8Q2N5 A0A0K8TM01 F4WH88 A0A139WF40 A0A023F813 A0A336MAA2 U5EXA1 A0A158NPD5 A0A212EH02 A0A0V0G8W0 E2BL25 A0A224XY42 A0A069DPU1 A0A151X651 A0A0P4VSW7 T1I3E0 A0A0A9WGX2 A0A146L853 A0A2H1VN45 A0A2R7W2U1 A0A088AH49 A0A162SKY8 A0A0P5M9Q6 A0A3L8DYF1 A0A1B6DE18 A0A2A3ET45 A0A2S2P754 A0A026WNY9 A0A2H8TPU4 A0A1B6JBA2 A0A310SJT8 A0A1L8E8I8 A0A1B6F8L4 A0A0M8ZYZ7 A0A1B6LWE2 A0A2S2QT32 A0A0P6BVY1
A0A0P8Y5U0 A0A0Q9W9H4 A0A182PTD8 A0A182RR81 A0A1S4H2D3 A0A182V6J5 A0A182XJK2 A0A182KTK3 A0A182HRI0 A0A2M4C174 A0A182YMS5 A0A182JTT1 A0A2M4C136 W5JSH0 A0A2M3Z7N2 A0A2M4AVX9 A0A0R3NSN0 B4G8J0 A0A1Q3FE64 A0A182U7I3 A0A1A9VJC2 A0A1W4V9D9 A0A182NAS6 B0WVS2 A0A182F3W1 A0A1L8DRT2 A0A182W906 B4JB54 D3TRV4 B4I5M1 B4Q8Z6 A0A139WF38 A0A0R1DPC5 A0A0Q5VUJ7 A0A182M460 A0A023EHV2 Q9VJ38 A0A0Q9X221 A0A0Q9X3E8 A0A0K8W6L9 J3JY52 W8C7W6 A0A0A1X9F2 Q17ET7 A0A1A9Y5N8 A0A240PLE7 Q7Q191 A0A1W4WU47 E2AXI1 A0A0L0BZT8 A0A1L8ED45 A0A1Y1MJK0 A0A1L8EDC2 A0A1B0DH47 A0A067RAN4 A0A0M5J9G6 A0A1I8MKJ8 A0A1I8Q2N5 A0A0K8TM01 F4WH88 A0A139WF40 A0A023F813 A0A336MAA2 U5EXA1 A0A158NPD5 A0A212EH02 A0A0V0G8W0 E2BL25 A0A224XY42 A0A069DPU1 A0A151X651 A0A0P4VSW7 T1I3E0 A0A0A9WGX2 A0A146L853 A0A2H1VN45 A0A2R7W2U1 A0A088AH49 A0A162SKY8 A0A0P5M9Q6 A0A3L8DYF1 A0A1B6DE18 A0A2A3ET45 A0A2S2P754 A0A026WNY9 A0A2H8TPU4 A0A1B6JBA2 A0A310SJT8 A0A1L8E8I8 A0A1B6F8L4 A0A0M8ZYZ7 A0A1B6LWE2 A0A2S2QT32 A0A0P6BVY1
Pubmed
19121390
26354079
17994087
12364791
20966253
25244985
+ More
20920257 23761445 15632085 20353571 22936249 18362917 19820115 17550304 24945155 10731132 12537572 22516182 23537049 24495485 25830018 17510324 20798317 26108605 28004739 24845553 25315136 26369729 21719571 25474469 21347285 22118469 26334808 27129103 25401762 26823975 30249741 24508170
20920257 23761445 15632085 20353571 22936249 18362917 19820115 17550304 24945155 10731132 12537572 22516182 23537049 24495485 25830018 17510324 20798317 26108605 28004739 24845553 25315136 26369729 21719571 25474469 21347285 22118469 26334808 27129103 25401762 26823975 30249741 24508170
EMBL
BABH01021632
BABH01021633
BABH01021634
BABH01021635
NWSH01004210
PCG65358.1
+ More
KQ460772 KPJ12491.1 KQ459185 KPJ03276.1 ODYU01002268 SOQ39527.1 AXCN02000660 CH902624 KPU74502.1 CH940649 KRF81411.1 AAAB01008980 APCN01001706 GGFJ01009873 MBW59014.1 GGFJ01009872 MBW59013.1 ADMH02000223 ETN67322.1 GGFM01003764 MBW24515.1 GGFK01011609 MBW44930.1 CH379060 KRT03948.1 CH479180 EDW28670.1 GFDL01009208 JAV25837.1 DS232131 EDS35679.1 GFDF01004886 JAV09198.1 CH916368 EDW03946.1 EZ424156 ADD20432.1 CH480822 EDW55677.1 CM000361 CM002910 EDX05428.1 KMY90892.1 KQ971354 KYB26481.1 CM000158 KRJ99130.1 CH954179 KQS61915.1 AXCM01000771 GAPW01004946 JAC08652.1 AE014134 BT053702 CH963852 KRF98263.1 CH933807 KRG02566.1 GDHF01005481 JAI46833.1 APGK01035291 BT128176 KB740923 KB632333 AEE63137.1 ENN78247.1 ERL92678.1 GAMC01000287 JAC06269.1 GBXI01006721 JAD07571.1 CH477279 EAT45015.1 EAA14080.4 GL443583 EFN61866.1 JRES01001109 KNC25501.1 GFDG01002227 JAV16572.1 GEZM01033892 JAV84066.1 GFDG01002226 JAV16573.1 AJVK01060700 KK852632 KDR19814.1 CP012523 ALC39981.1 GDAI01002219 JAI15384.1 GL888153 EGI66454.1 KYB26479.1 GBBI01001343 JAC17369.1 UFQT01000618 SSX25819.1 GANO01001040 JAB58831.1 ADTU01022391 AGBW02014995 OWR40756.1 GECL01001659 JAP04465.1 GL448902 EFN83603.1 GFTR01002944 JAW13482.1 GBGD01003183 JAC85706.1 KQ982482 KYQ55861.1 GDKW01000900 JAI55695.1 ACPB03010447 GBHO01045181 GBHO01037826 GBRD01001060 GBRD01001059 JAF98422.1 JAG05778.1 JAG64761.1 GDHC01014055 JAQ04574.1 ODYU01003454 SOQ42255.1 KK854251 PTY13738.1 LRGB01000024 KZS21405.1 GDIQ01161341 JAK90384.1 QOIP01000003 RLU24979.1 GEDC01031130 GEDC01026628 GEDC01013402 JAS06168.1 JAS10670.1 JAS23896.1 KZ288189 PBC34684.1 GGMR01012419 MBY25038.1 KK107139 EZA57653.1 GFXV01004116 MBW15921.1 GECU01011349 JAS96357.1 KQ763877 OAD54652.1 GFDG01003719 JAV15080.1 GECZ01023190 JAS46579.1 KQ435794 KOX73980.1 GEBQ01029587 GEBQ01011991 JAT10390.1 JAT27986.1 GGMS01011477 MBY80680.1 GDIP01009899 JAM93816.1
KQ460772 KPJ12491.1 KQ459185 KPJ03276.1 ODYU01002268 SOQ39527.1 AXCN02000660 CH902624 KPU74502.1 CH940649 KRF81411.1 AAAB01008980 APCN01001706 GGFJ01009873 MBW59014.1 GGFJ01009872 MBW59013.1 ADMH02000223 ETN67322.1 GGFM01003764 MBW24515.1 GGFK01011609 MBW44930.1 CH379060 KRT03948.1 CH479180 EDW28670.1 GFDL01009208 JAV25837.1 DS232131 EDS35679.1 GFDF01004886 JAV09198.1 CH916368 EDW03946.1 EZ424156 ADD20432.1 CH480822 EDW55677.1 CM000361 CM002910 EDX05428.1 KMY90892.1 KQ971354 KYB26481.1 CM000158 KRJ99130.1 CH954179 KQS61915.1 AXCM01000771 GAPW01004946 JAC08652.1 AE014134 BT053702 CH963852 KRF98263.1 CH933807 KRG02566.1 GDHF01005481 JAI46833.1 APGK01035291 BT128176 KB740923 KB632333 AEE63137.1 ENN78247.1 ERL92678.1 GAMC01000287 JAC06269.1 GBXI01006721 JAD07571.1 CH477279 EAT45015.1 EAA14080.4 GL443583 EFN61866.1 JRES01001109 KNC25501.1 GFDG01002227 JAV16572.1 GEZM01033892 JAV84066.1 GFDG01002226 JAV16573.1 AJVK01060700 KK852632 KDR19814.1 CP012523 ALC39981.1 GDAI01002219 JAI15384.1 GL888153 EGI66454.1 KYB26479.1 GBBI01001343 JAC17369.1 UFQT01000618 SSX25819.1 GANO01001040 JAB58831.1 ADTU01022391 AGBW02014995 OWR40756.1 GECL01001659 JAP04465.1 GL448902 EFN83603.1 GFTR01002944 JAW13482.1 GBGD01003183 JAC85706.1 KQ982482 KYQ55861.1 GDKW01000900 JAI55695.1 ACPB03010447 GBHO01045181 GBHO01037826 GBRD01001060 GBRD01001059 JAF98422.1 JAG05778.1 JAG64761.1 GDHC01014055 JAQ04574.1 ODYU01003454 SOQ42255.1 KK854251 PTY13738.1 LRGB01000024 KZS21405.1 GDIQ01161341 JAK90384.1 QOIP01000003 RLU24979.1 GEDC01031130 GEDC01026628 GEDC01013402 JAS06168.1 JAS10670.1 JAS23896.1 KZ288189 PBC34684.1 GGMR01012419 MBY25038.1 KK107139 EZA57653.1 GFXV01004116 MBW15921.1 GECU01011349 JAS96357.1 KQ763877 OAD54652.1 GFDG01003719 JAV15080.1 GECZ01023190 JAS46579.1 KQ435794 KOX73980.1 GEBQ01029587 GEBQ01011991 JAT10390.1 JAT27986.1 GGMS01011477 MBY80680.1 GDIP01009899 JAM93816.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000075886
UP000007801
+ More
UP000008792 UP000075885 UP000075900 UP000075903 UP000076407 UP000075882 UP000075840 UP000076408 UP000075881 UP000000673 UP000001819 UP000008744 UP000075902 UP000078200 UP000192221 UP000075884 UP000002320 UP000069272 UP000075920 UP000001070 UP000001292 UP000000304 UP000007266 UP000002282 UP000008711 UP000075883 UP000000803 UP000007798 UP000009192 UP000019118 UP000030742 UP000008820 UP000092443 UP000075880 UP000007062 UP000192223 UP000000311 UP000037069 UP000092462 UP000027135 UP000092553 UP000095301 UP000095300 UP000007755 UP000005205 UP000007151 UP000008237 UP000075809 UP000015103 UP000005203 UP000076858 UP000279307 UP000242457 UP000053097 UP000053105
UP000008792 UP000075885 UP000075900 UP000075903 UP000076407 UP000075882 UP000075840 UP000076408 UP000075881 UP000000673 UP000001819 UP000008744 UP000075902 UP000078200 UP000192221 UP000075884 UP000002320 UP000069272 UP000075920 UP000001070 UP000001292 UP000000304 UP000007266 UP000002282 UP000008711 UP000075883 UP000000803 UP000007798 UP000009192 UP000019118 UP000030742 UP000008820 UP000092443 UP000075880 UP000007062 UP000192223 UP000000311 UP000037069 UP000092462 UP000027135 UP000092553 UP000095301 UP000095300 UP000007755 UP000005205 UP000007151 UP000008237 UP000075809 UP000015103 UP000005203 UP000076858 UP000279307 UP000242457 UP000053097 UP000053105
Pfam
Interpro
IPR034015
M1_LTA4H
+ More
IPR001930 Peptidase_M1
IPR036899 Ribosomal_L13_sf
IPR015211 Peptidase_M1_C
IPR027268 Peptidase_M4/M1_CTD_sf
IPR038502 M1_LTA-4_hydro/amino_C_sf
IPR016024 ARM-type_fold
IPR014782 Peptidase_M1_dom
IPR042097 Aminopeptidase_N-like_N
IPR005822 Ribosomal_L13
IPR005823 Ribosomal_L13_bac-type
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR035899 DBL_dom_sf
IPR041020 PH_16
IPR001478 PDZ
IPR010796 B9_dom
IPR001849 PH_domain
IPR000219 DH-domain
IPR016137 RGS
IPR036305 RGS_sf
IPR015212 RGS-like_dom
IPR011677 DUF1619
IPR036034 PDZ_sf
IPR011993 PH-like_dom_sf
IPR002219 PE/DAG-bd
IPR001930 Peptidase_M1
IPR036899 Ribosomal_L13_sf
IPR015211 Peptidase_M1_C
IPR027268 Peptidase_M4/M1_CTD_sf
IPR038502 M1_LTA-4_hydro/amino_C_sf
IPR016024 ARM-type_fold
IPR014782 Peptidase_M1_dom
IPR042097 Aminopeptidase_N-like_N
IPR005822 Ribosomal_L13
IPR005823 Ribosomal_L13_bac-type
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR035899 DBL_dom_sf
IPR041020 PH_16
IPR001478 PDZ
IPR010796 B9_dom
IPR001849 PH_domain
IPR000219 DH-domain
IPR016137 RGS
IPR036305 RGS_sf
IPR015212 RGS-like_dom
IPR011677 DUF1619
IPR036034 PDZ_sf
IPR011993 PH-like_dom_sf
IPR002219 PE/DAG-bd
SUPFAM
ProteinModelPortal
H9J3M5
A0A2A4J110
A0A0N0PBZ0
A0A194QIJ7
A0A2H1VFC0
A0A182QKX5
+ More
A0A0P8Y5U0 A0A0Q9W9H4 A0A182PTD8 A0A182RR81 A0A1S4H2D3 A0A182V6J5 A0A182XJK2 A0A182KTK3 A0A182HRI0 A0A2M4C174 A0A182YMS5 A0A182JTT1 A0A2M4C136 W5JSH0 A0A2M3Z7N2 A0A2M4AVX9 A0A0R3NSN0 B4G8J0 A0A1Q3FE64 A0A182U7I3 A0A1A9VJC2 A0A1W4V9D9 A0A182NAS6 B0WVS2 A0A182F3W1 A0A1L8DRT2 A0A182W906 B4JB54 D3TRV4 B4I5M1 B4Q8Z6 A0A139WF38 A0A0R1DPC5 A0A0Q5VUJ7 A0A182M460 A0A023EHV2 Q9VJ38 A0A0Q9X221 A0A0Q9X3E8 A0A0K8W6L9 J3JY52 W8C7W6 A0A0A1X9F2 Q17ET7 A0A1A9Y5N8 A0A240PLE7 Q7Q191 A0A1W4WU47 E2AXI1 A0A0L0BZT8 A0A1L8ED45 A0A1Y1MJK0 A0A1L8EDC2 A0A1B0DH47 A0A067RAN4 A0A0M5J9G6 A0A1I8MKJ8 A0A1I8Q2N5 A0A0K8TM01 F4WH88 A0A139WF40 A0A023F813 A0A336MAA2 U5EXA1 A0A158NPD5 A0A212EH02 A0A0V0G8W0 E2BL25 A0A224XY42 A0A069DPU1 A0A151X651 A0A0P4VSW7 T1I3E0 A0A0A9WGX2 A0A146L853 A0A2H1VN45 A0A2R7W2U1 A0A088AH49 A0A162SKY8 A0A0P5M9Q6 A0A3L8DYF1 A0A1B6DE18 A0A2A3ET45 A0A2S2P754 A0A026WNY9 A0A2H8TPU4 A0A1B6JBA2 A0A310SJT8 A0A1L8E8I8 A0A1B6F8L4 A0A0M8ZYZ7 A0A1B6LWE2 A0A2S2QT32 A0A0P6BVY1
A0A0P8Y5U0 A0A0Q9W9H4 A0A182PTD8 A0A182RR81 A0A1S4H2D3 A0A182V6J5 A0A182XJK2 A0A182KTK3 A0A182HRI0 A0A2M4C174 A0A182YMS5 A0A182JTT1 A0A2M4C136 W5JSH0 A0A2M3Z7N2 A0A2M4AVX9 A0A0R3NSN0 B4G8J0 A0A1Q3FE64 A0A182U7I3 A0A1A9VJC2 A0A1W4V9D9 A0A182NAS6 B0WVS2 A0A182F3W1 A0A1L8DRT2 A0A182W906 B4JB54 D3TRV4 B4I5M1 B4Q8Z6 A0A139WF38 A0A0R1DPC5 A0A0Q5VUJ7 A0A182M460 A0A023EHV2 Q9VJ38 A0A0Q9X221 A0A0Q9X3E8 A0A0K8W6L9 J3JY52 W8C7W6 A0A0A1X9F2 Q17ET7 A0A1A9Y5N8 A0A240PLE7 Q7Q191 A0A1W4WU47 E2AXI1 A0A0L0BZT8 A0A1L8ED45 A0A1Y1MJK0 A0A1L8EDC2 A0A1B0DH47 A0A067RAN4 A0A0M5J9G6 A0A1I8MKJ8 A0A1I8Q2N5 A0A0K8TM01 F4WH88 A0A139WF40 A0A023F813 A0A336MAA2 U5EXA1 A0A158NPD5 A0A212EH02 A0A0V0G8W0 E2BL25 A0A224XY42 A0A069DPU1 A0A151X651 A0A0P4VSW7 T1I3E0 A0A0A9WGX2 A0A146L853 A0A2H1VN45 A0A2R7W2U1 A0A088AH49 A0A162SKY8 A0A0P5M9Q6 A0A3L8DYF1 A0A1B6DE18 A0A2A3ET45 A0A2S2P754 A0A026WNY9 A0A2H8TPU4 A0A1B6JBA2 A0A310SJT8 A0A1L8E8I8 A0A1B6F8L4 A0A0M8ZYZ7 A0A1B6LWE2 A0A2S2QT32 A0A0P6BVY1
PDB
6GB2
E-value=2.93991e-44,
Score=445
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion
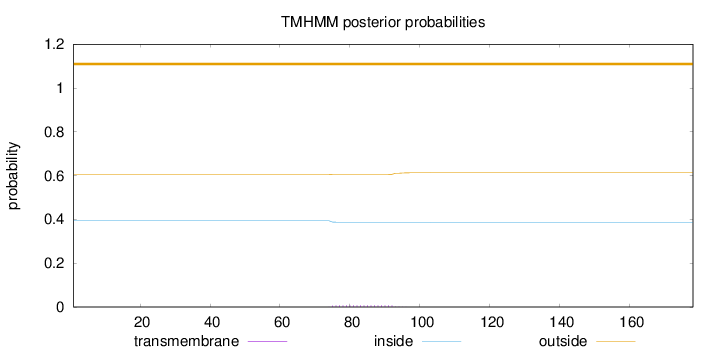
Length:
178
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13907
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.39414
outside
1 - 178
Population Genetic Test Statistics
Pi
150.254457
Theta
168.444029
Tajima's D
0.077228
CLR
0.090059
CSRT
0.389230538473076
Interpretation
Possibly Positive selection
