Gene
KWMTBOMO11613 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004112
Annotation
leukotriene_A-4_hydrolase_[Bombyx_mori]
Full name
Leukotriene A(4) hydrolase
Location in the cell
Cytoplasmic Reliability : 2.788
Sequence
CDS
ATGGGTGCTTTCAGTCCTCTGGATCCGTCTTCCTTTTCGCGGCCTGAACAAGCTGTAATAAAACACGTGACTTTATCGTTGAACGTGGATTTTGAAAACAAAGTCTTAAACGGATCCGCCACTCTCGACGTTGACGTGCTCCAAGATATCGGAGACGTGGTCCTTGATTCGAGTGAACTGACAATAGAATCAATAGAACTAGACGGTGCTCAATTGACGTACAAATTAGATGACCCAGTGCCGAACTATGGCTCCAAATTGACTATACAATTGCCGAAACGAGCCTCAAGTGGCGATAAGTTAAAAATTAAAATTAAGTACACAACGTCCCCGTCCGCGACTGCGTTACAATGGTTACAGCCAGCTCAAACTTCGGGGAAGAAACACCCTTATCTATTCAGCCAGTGTCAGCCGATTCATGCCCGTTCGATTCTTCCGTGTCAAGACACTCCATTTGTAAAGTTCACTTATGACGCTGAGGTCACGGCGCCCGAGGAGTTCACAGTACTGATGAGCGCGCTCAGAGGCGAGAGTCGCAGCACTAAGACGACCTTCAATCAGCCGATGCCTTTGCCGTCGTATCTTTTGGCCATTGCGGTCGGCGTGTTGGAGCACAGGACTCTAGGACCCAGGTCTAAAGTTTGGTCCGAGAAAGAGGAGATCGAACGTAGCGCTTGGGAGTTCGCTGAGGCCGAGAAATATCTGCAAGCCGCCGAAAGACTCTGCGGCGCTTACGAATGGAAACAGTACGACTTGCTGGTGCTGCCACCTTCCTTCCCTTACGGGGGGATGGAGAACCCGTGCCTTACTTTTGTAACGCCCACTTTGTTGGCTGGTGATAGAAGTCAAGCAGACGTGTTGATCCACGAGATCATGCACAGTTGGACCGGTAACCTCGTCACGAATAGAAATTTTGAACATTTCTGGCTAAACGAGGGCTTCACTGTATTCCTGGAGAGAAAAGTCAAAGCTTCTTTGATAGAAGATCCAATTGAAGCCAAGAAAAGCAGAGACTTTCATAGTATACTCGGACTGGAGGAGTTAAGTGAAGCGGTGGTAAGTCAGTTAGGCGTTGACAATCCTCTGACCCGGTTGGTACCCGACCTCCATGACGTGCATCCTGACGATGCATTCTCCACGGTTCCCTACGAAAAGGGCTCGCTGTTTTTGCGATACCTTGAGGACTTAATAGGAGGACCAGAAGTATTCGATGATTTCCTACGATCGTACCTGAAGAACTTCAGTCGGAAATCTCTTGACACGGACGAGTTCAAGGCCTATCTGTTTAACTATTTCCAAGACAATGAAAAATTGAAGAGCGTTGATTGGGAAACATGGTTGTACAAGAGCGGTATGCCCCCCGTCATACCAGACTACGACACTTCAATGACGAAACCGATAACGGCTCTCGTTGCGAAGATCAACCAGCGCAACGCGTCGCTGATATCGTATCAAGACGTGCAAACGTTTACTCCTCATCAAATAATGATATTCCTGCAAGAACTGATAAAACATCCTCCACTGCCGCTAGACATTCTCCACACTCTTGGCGCAGAGTATAAAATCGATAGTAGCAAGAATACCGAGATTTTGTATAGATGGTTGCGGCTGTGCCTTAGAAGCAAAGATCAATCGATCTTGGGCGAAGTGTTCGATTTTGTGAACCACATAGGACGGATGCGGTACGTCCGTCCACTGTATAGAGACCTCTACGATTGGGAAGACGTTAGAAATCAAGCAATAGAAAACTTCCTCAAAAACGAACAATACATGATGCACGTGTGTGCTCATACTGTGAAAAAAGATTTACATTTAATTGACTAG
Protein
MGAFSPLDPSSFSRPEQAVIKHVTLSLNVDFENKVLNGSATLDVDVLQDIGDVVLDSSELTIESIELDGAQLTYKLDDPVPNYGSKLTIQLPKRASSGDKLKIKIKYTTSPSATALQWLQPAQTSGKKHPYLFSQCQPIHARSILPCQDTPFVKFTYDAEVTAPEEFTVLMSALRGESRSTKTTFNQPMPLPSYLLAIAVGVLEHRTLGPRSKVWSEKEEIERSAWEFAEAEKYLQAAERLCGAYEWKQYDLLVLPPSFPYGGMENPCLTFVTPTLLAGDRSQADVLIHEIMHSWTGNLVTNRNFEHFWLNEGFTVFLERKVKASLIEDPIEAKKSRDFHSILGLEELSEAVVSQLGVDNPLTRLVPDLHDVHPDDAFSTVPYEKGSLFLRYLEDLIGGPEVFDDFLRSYLKNFSRKSLDTDEFKAYLFNYFQDNEKLKSVDWETWLYKSGMPPVIPDYDTSMTKPITALVAKINQRNASLISYQDVQTFTPHQIMIFLQELIKHPPLPLDILHTLGAEYKIDSSKNTEILYRWLRLCLRSKDQSILGEVFDFVNHIGRMRYVRPLYRDLYDWEDVRNQAIENFLKNEQYMMHVCAHTVKKDLHLID
Summary
Catalytic Activity
H2O + leukotriene A4 = leukotriene B4
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Feature
chain Leukotriene A(4) hydrolase
Uniprot
H9J3M5
Q1HPZ6
A0A2A4J1R9
A0A2A4J028
A0A2A4IZL0
A0A2A4J1U2
+ More
A0A2A4J0U5 A0A212EGY7 A0A194QEA7 S4PDP9 A0A0N1INX3 I4DMU9 E2AJV4 A0A3L8DEW0 A0A026WSW2 A0A067R2V8 D6WQA3 N6TK18 A0A0J7NQV8 A0A2J7RI89 K7IPJ4 A0A232F4W0 A0A087SXZ5 A0A1S3HTK7 A0A0P4WA18 A0A1B6EZ69 A0A0P5HVT5 A0A0P5HTE4 A0A0P6I2V5 A0A162P439 A0A0P6D086 A0A1W4WJA4 A0A034VLU1 A0A0L0BZJ4 A0A2H8TMS3 A0A0K8V4Y7 A0A0P6F148 A0A1I8MPT4 A0A1I8MPT3 A0A0K8WE97 A0A1Y1MLJ2 T1PAM3 A0A0A1XDU2 A0A0K8V3D7 W8BRL6 Q17EU2 A0A1S4F5A1 A0A293MVW2 A0A1Q3FAS5 U5EZH8 E9G305 A0A336KHS5 A0A210PJC8 A0A0P6EKM3 A0A2M4AIT2 A0A2M4BHC6 A0A2M4BHM9 A0A1Q3FB79 A0A182F3W0 A0A084W8N2 W5JVF8 B4JB55 B4KHC6 A0A182ILY2 E2BTZ7 A0A1L8EA25 T1JM78 A0A0Q9XER5 A0A3B0J9M7 A0A2J7RI85 A0A1Z5L5Q1 A0A182RR80 J9JSZ2 B4LU93 A0A0P5I5C9 A0A1L8EA35 A0A182JTT2 A0A1L8EAE0 A0A1L8E9Y8 A0A182TNS2 B0WVS1 A0A3B0JE93 B5DI51 A0A1I8Q2R3 A0A0Q9WLT2 A0A0R3NXG4 A0A182HRI1 A0A182XJK3 U4UFC5 A0A1I8Q2N7 A0A182VM67 B4G8I9 Q7Q192 A0A1S4H3A8 A0A182KTJ7 A0A0K2SWE2 B4MTS5 A0A023EVM8 A0A0M4EGR4 G3MF29
A0A2A4J0U5 A0A212EGY7 A0A194QEA7 S4PDP9 A0A0N1INX3 I4DMU9 E2AJV4 A0A3L8DEW0 A0A026WSW2 A0A067R2V8 D6WQA3 N6TK18 A0A0J7NQV8 A0A2J7RI89 K7IPJ4 A0A232F4W0 A0A087SXZ5 A0A1S3HTK7 A0A0P4WA18 A0A1B6EZ69 A0A0P5HVT5 A0A0P5HTE4 A0A0P6I2V5 A0A162P439 A0A0P6D086 A0A1W4WJA4 A0A034VLU1 A0A0L0BZJ4 A0A2H8TMS3 A0A0K8V4Y7 A0A0P6F148 A0A1I8MPT4 A0A1I8MPT3 A0A0K8WE97 A0A1Y1MLJ2 T1PAM3 A0A0A1XDU2 A0A0K8V3D7 W8BRL6 Q17EU2 A0A1S4F5A1 A0A293MVW2 A0A1Q3FAS5 U5EZH8 E9G305 A0A336KHS5 A0A210PJC8 A0A0P6EKM3 A0A2M4AIT2 A0A2M4BHC6 A0A2M4BHM9 A0A1Q3FB79 A0A182F3W0 A0A084W8N2 W5JVF8 B4JB55 B4KHC6 A0A182ILY2 E2BTZ7 A0A1L8EA25 T1JM78 A0A0Q9XER5 A0A3B0J9M7 A0A2J7RI85 A0A1Z5L5Q1 A0A182RR80 J9JSZ2 B4LU93 A0A0P5I5C9 A0A1L8EA35 A0A182JTT2 A0A1L8EAE0 A0A1L8E9Y8 A0A182TNS2 B0WVS1 A0A3B0JE93 B5DI51 A0A1I8Q2R3 A0A0Q9WLT2 A0A0R3NXG4 A0A182HRI1 A0A182XJK3 U4UFC5 A0A1I8Q2N7 A0A182VM67 B4G8I9 Q7Q192 A0A1S4H3A8 A0A182KTJ7 A0A0K2SWE2 B4MTS5 A0A023EVM8 A0A0M4EGR4 G3MF29
EC Number
3.3.2.6
Pubmed
19121390
22118469
26354079
23622113
22651552
20798317
+ More
30249741 24508170 24845553 18362917 19820115 23537049 20075255 28648823 25348373 26108605 25315136 28004739 25830018 24495485 17510324 21292972 28812685 24438588 20920257 23761445 17994087 28528879 18057021 15632085 12364791 20966253 24945155 22216098
30249741 24508170 24845553 18362917 19820115 23537049 20075255 28648823 25348373 26108605 25315136 28004739 25830018 24495485 17510324 21292972 28812685 24438588 20920257 23761445 17994087 28528879 18057021 15632085 12364791 20966253 24945155 22216098
EMBL
BABH01021632
BABH01021633
BABH01021634
BABH01021635
DQ443256
ABF51345.1
+ More
NWSH01004210 PCG65352.1 PCG65351.1 PCG65357.1 PCG65353.1 PCG65356.1 AGBW02014995 OWR40757.1 KQ459185 KPJ03280.1 GAIX01003576 JAA88984.1 KQ460772 KPJ12486.1 AK402617 BAM19239.1 GL440100 EFN66270.1 QOIP01000009 RLU18861.1 KK107119 EZA58766.1 KK852949 KDR13370.1 KQ971354 EFA07524.1 APGK01035291 APGK01035292 KB740923 ENN78248.1 LBMM01002417 KMQ94850.1 NEVH01003504 PNF40549.1 NNAY01001008 OXU25510.1 KK112471 KFM57734.1 GDRN01079561 JAI62381.1 GECZ01026423 JAS43346.1 GDIQ01224453 JAK27272.1 GDIQ01223199 JAK28526.1 GDIQ01026659 JAN68078.1 LRGB01000512 KZS18502.1 GDIP01007387 JAM96328.1 GAKP01016434 GAKP01016433 JAC42518.1 JRES01001109 KNC25502.1 GFXV01003669 MBW15474.1 GDHF01018312 JAI34002.1 GDIQ01063011 JAN31726.1 GDHF01033565 GDHF01016049 GDHF01006128 GDHF01002826 JAI18749.1 JAI36265.1 JAI46186.1 JAI49488.1 GEZM01033892 JAV84067.1 KA645821 AFP60450.1 GBXI01015035 GBXI01005659 JAC99256.1 JAD08633.1 GDHF01018877 JAI33437.1 GAMC01007042 JAB99513.1 CH477279 EAT45009.1 GFWV01019521 MAA44249.1 GFDL01010361 JAV24684.1 GANO01000771 JAB59100.1 GL732530 EFX86132.1 UFQS01000439 UFQT01000439 SSX03967.1 SSX24332.1 NEDP02076559 OWF36589.1 GDIQ01061218 JAN33519.1 GGFK01007321 MBW40642.1 GGFJ01003319 MBW52460.1 GGFJ01003320 MBW52461.1 GFDL01010238 JAV24807.1 ATLV01021496 KE525319 KFB46576.1 ADMH02000223 ETN67323.1 CH916368 EDW03947.1 CH933807 EDW11190.1 GL450531 EFN80896.1 GFDG01003212 JAV15587.1 JH431873 KRG02567.1 OUUW01000004 SPP78974.1 PNF40547.1 GFJQ02004586 JAW02384.1 ABLF02033844 CH940649 EDW64080.1 KRF81412.1 GDIQ01224454 JAK27271.1 GFDG01003211 JAV15588.1 GFDG01003210 JAV15589.1 GFDG01003209 JAV15590.1 DS232131 EDS35678.1 SPP78973.1 CH379060 EDY69989.1 KRF81413.1 KRT03949.1 APCN01001706 KB632333 ERL92679.1 CH479180 EDW28669.1 AAAB01008980 EAA14067.4 HACA01000708 CDW18069.1 CH963852 EDW75514.2 GAPW01000492 JAC13106.1 CP012523 ALC39980.1 JO840480 AEO32097.1
NWSH01004210 PCG65352.1 PCG65351.1 PCG65357.1 PCG65353.1 PCG65356.1 AGBW02014995 OWR40757.1 KQ459185 KPJ03280.1 GAIX01003576 JAA88984.1 KQ460772 KPJ12486.1 AK402617 BAM19239.1 GL440100 EFN66270.1 QOIP01000009 RLU18861.1 KK107119 EZA58766.1 KK852949 KDR13370.1 KQ971354 EFA07524.1 APGK01035291 APGK01035292 KB740923 ENN78248.1 LBMM01002417 KMQ94850.1 NEVH01003504 PNF40549.1 NNAY01001008 OXU25510.1 KK112471 KFM57734.1 GDRN01079561 JAI62381.1 GECZ01026423 JAS43346.1 GDIQ01224453 JAK27272.1 GDIQ01223199 JAK28526.1 GDIQ01026659 JAN68078.1 LRGB01000512 KZS18502.1 GDIP01007387 JAM96328.1 GAKP01016434 GAKP01016433 JAC42518.1 JRES01001109 KNC25502.1 GFXV01003669 MBW15474.1 GDHF01018312 JAI34002.1 GDIQ01063011 JAN31726.1 GDHF01033565 GDHF01016049 GDHF01006128 GDHF01002826 JAI18749.1 JAI36265.1 JAI46186.1 JAI49488.1 GEZM01033892 JAV84067.1 KA645821 AFP60450.1 GBXI01015035 GBXI01005659 JAC99256.1 JAD08633.1 GDHF01018877 JAI33437.1 GAMC01007042 JAB99513.1 CH477279 EAT45009.1 GFWV01019521 MAA44249.1 GFDL01010361 JAV24684.1 GANO01000771 JAB59100.1 GL732530 EFX86132.1 UFQS01000439 UFQT01000439 SSX03967.1 SSX24332.1 NEDP02076559 OWF36589.1 GDIQ01061218 JAN33519.1 GGFK01007321 MBW40642.1 GGFJ01003319 MBW52460.1 GGFJ01003320 MBW52461.1 GFDL01010238 JAV24807.1 ATLV01021496 KE525319 KFB46576.1 ADMH02000223 ETN67323.1 CH916368 EDW03947.1 CH933807 EDW11190.1 GL450531 EFN80896.1 GFDG01003212 JAV15587.1 JH431873 KRG02567.1 OUUW01000004 SPP78974.1 PNF40547.1 GFJQ02004586 JAW02384.1 ABLF02033844 CH940649 EDW64080.1 KRF81412.1 GDIQ01224454 JAK27271.1 GFDG01003211 JAV15588.1 GFDG01003210 JAV15589.1 GFDG01003209 JAV15590.1 DS232131 EDS35678.1 SPP78973.1 CH379060 EDY69989.1 KRF81413.1 KRT03949.1 APCN01001706 KB632333 ERL92679.1 CH479180 EDW28669.1 AAAB01008980 EAA14067.4 HACA01000708 CDW18069.1 CH963852 EDW75514.2 GAPW01000492 JAC13106.1 CP012523 ALC39980.1 JO840480 AEO32097.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000000311
+ More
UP000279307 UP000053097 UP000027135 UP000007266 UP000019118 UP000036403 UP000235965 UP000002358 UP000215335 UP000054359 UP000085678 UP000076858 UP000192223 UP000037069 UP000095301 UP000008820 UP000000305 UP000242188 UP000069272 UP000030765 UP000000673 UP000001070 UP000009192 UP000075880 UP000008237 UP000268350 UP000075900 UP000007819 UP000008792 UP000075881 UP000075902 UP000002320 UP000001819 UP000095300 UP000075840 UP000076407 UP000030742 UP000075903 UP000008744 UP000007062 UP000075882 UP000007798 UP000092553
UP000279307 UP000053097 UP000027135 UP000007266 UP000019118 UP000036403 UP000235965 UP000002358 UP000215335 UP000054359 UP000085678 UP000076858 UP000192223 UP000037069 UP000095301 UP000008820 UP000000305 UP000242188 UP000069272 UP000030765 UP000000673 UP000001070 UP000009192 UP000075880 UP000008237 UP000268350 UP000075900 UP000007819 UP000008792 UP000075881 UP000075902 UP000002320 UP000001819 UP000095300 UP000075840 UP000076407 UP000030742 UP000075903 UP000008744 UP000007062 UP000075882 UP000007798 UP000092553
Interpro
ProteinModelPortal
H9J3M5
Q1HPZ6
A0A2A4J1R9
A0A2A4J028
A0A2A4IZL0
A0A2A4J1U2
+ More
A0A2A4J0U5 A0A212EGY7 A0A194QEA7 S4PDP9 A0A0N1INX3 I4DMU9 E2AJV4 A0A3L8DEW0 A0A026WSW2 A0A067R2V8 D6WQA3 N6TK18 A0A0J7NQV8 A0A2J7RI89 K7IPJ4 A0A232F4W0 A0A087SXZ5 A0A1S3HTK7 A0A0P4WA18 A0A1B6EZ69 A0A0P5HVT5 A0A0P5HTE4 A0A0P6I2V5 A0A162P439 A0A0P6D086 A0A1W4WJA4 A0A034VLU1 A0A0L0BZJ4 A0A2H8TMS3 A0A0K8V4Y7 A0A0P6F148 A0A1I8MPT4 A0A1I8MPT3 A0A0K8WE97 A0A1Y1MLJ2 T1PAM3 A0A0A1XDU2 A0A0K8V3D7 W8BRL6 Q17EU2 A0A1S4F5A1 A0A293MVW2 A0A1Q3FAS5 U5EZH8 E9G305 A0A336KHS5 A0A210PJC8 A0A0P6EKM3 A0A2M4AIT2 A0A2M4BHC6 A0A2M4BHM9 A0A1Q3FB79 A0A182F3W0 A0A084W8N2 W5JVF8 B4JB55 B4KHC6 A0A182ILY2 E2BTZ7 A0A1L8EA25 T1JM78 A0A0Q9XER5 A0A3B0J9M7 A0A2J7RI85 A0A1Z5L5Q1 A0A182RR80 J9JSZ2 B4LU93 A0A0P5I5C9 A0A1L8EA35 A0A182JTT2 A0A1L8EAE0 A0A1L8E9Y8 A0A182TNS2 B0WVS1 A0A3B0JE93 B5DI51 A0A1I8Q2R3 A0A0Q9WLT2 A0A0R3NXG4 A0A182HRI1 A0A182XJK3 U4UFC5 A0A1I8Q2N7 A0A182VM67 B4G8I9 Q7Q192 A0A1S4H3A8 A0A182KTJ7 A0A0K2SWE2 B4MTS5 A0A023EVM8 A0A0M4EGR4 G3MF29
A0A2A4J0U5 A0A212EGY7 A0A194QEA7 S4PDP9 A0A0N1INX3 I4DMU9 E2AJV4 A0A3L8DEW0 A0A026WSW2 A0A067R2V8 D6WQA3 N6TK18 A0A0J7NQV8 A0A2J7RI89 K7IPJ4 A0A232F4W0 A0A087SXZ5 A0A1S3HTK7 A0A0P4WA18 A0A1B6EZ69 A0A0P5HVT5 A0A0P5HTE4 A0A0P6I2V5 A0A162P439 A0A0P6D086 A0A1W4WJA4 A0A034VLU1 A0A0L0BZJ4 A0A2H8TMS3 A0A0K8V4Y7 A0A0P6F148 A0A1I8MPT4 A0A1I8MPT3 A0A0K8WE97 A0A1Y1MLJ2 T1PAM3 A0A0A1XDU2 A0A0K8V3D7 W8BRL6 Q17EU2 A0A1S4F5A1 A0A293MVW2 A0A1Q3FAS5 U5EZH8 E9G305 A0A336KHS5 A0A210PJC8 A0A0P6EKM3 A0A2M4AIT2 A0A2M4BHC6 A0A2M4BHM9 A0A1Q3FB79 A0A182F3W0 A0A084W8N2 W5JVF8 B4JB55 B4KHC6 A0A182ILY2 E2BTZ7 A0A1L8EA25 T1JM78 A0A0Q9XER5 A0A3B0J9M7 A0A2J7RI85 A0A1Z5L5Q1 A0A182RR80 J9JSZ2 B4LU93 A0A0P5I5C9 A0A1L8EA35 A0A182JTT2 A0A1L8EAE0 A0A1L8E9Y8 A0A182TNS2 B0WVS1 A0A3B0JE93 B5DI51 A0A1I8Q2R3 A0A0Q9WLT2 A0A0R3NXG4 A0A182HRI1 A0A182XJK3 U4UFC5 A0A1I8Q2N7 A0A182VM67 B4G8I9 Q7Q192 A0A1S4H3A8 A0A182KTJ7 A0A0K2SWE2 B4MTS5 A0A023EVM8 A0A0M4EGR4 G3MF29
PDB
4GAA
E-value=3.07906e-163,
Score=1478
Ontologies
PATHWAY
GO
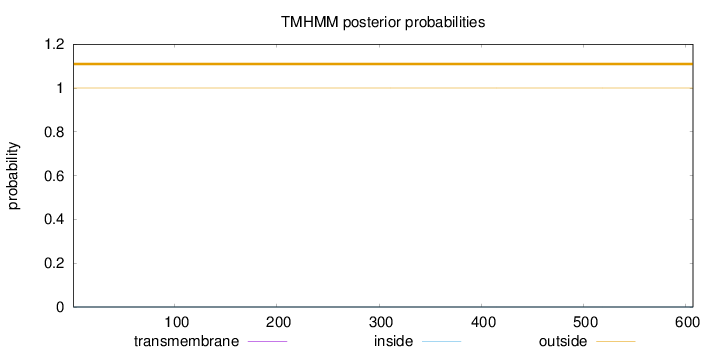
Topology
Subcellular location
Cytoplasm
Length:
607
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00175
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00011
outside
1 - 607
Population Genetic Test Statistics
Pi
221.79814
Theta
161.314134
Tajima's D
1.194034
CLR
0.021136
CSRT
0.713814309284536
Interpretation
Uncertain
