Gene
KWMTBOMO11612
Annotation
PREDICTED:_neuralized-like_protein_2_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.068
Sequence
CDS
ATGCTATCGAGATTCCATCATTATCATGGCTCGAATATTATATTGTTCGAAGAAAATACAGTTGCCTACAGAAAAGCTAGTTTTGCACACGGCTTGACTTTTAGTGAAAAACCCCTTCTACCTGGTGAAATATTTCTAGTAGAAATTGAAAAAACCGAAAGAGGATGGAGTGGTCATATGCGGCTCGGGTTAACACTTCTTGAGCCACAAGCAGCGGCTCTGGGTGACAAAGGATTGCCTCAGTATGCGCTACCAGATCTCGCTAACAATGGGCGATGGGAAAACGAGCCGCGGGTTTCGGTCCTAGGCGATGGTCATGTAGTAAAAACGCCACGTGGTTACATATCTAAGATGATTTTAAAGCCACAGACTGTCTCACAGAATGGAACACCCCAAGGTATTTTACCTATGGACGCTGGCAGCCGTATTGGAGTAATGTTTGTTCCTTGTCCTAAACTTAATAAAAATGACCAAGACTTAGCTGAAATGCACTTTATAATTAATGGTGAAGATCAAGGCCCTTGTACTAAATCGATACCCTACACAAGAGGTCTTTTACATGCAGTCGTTGATGTCTATGGTACAACAAAACAAGTGAAGATTGTACAACTTTATGGAGTGAAATCTCTTCAGATTATTTGCAGGGATGCAATACTGCAGAATGTGAAAAATGGTTCGGTGAAATCTTTACCCTTACCTAAAAGCCTAAAAGACTTTTTGTTATCATAA
Protein
MLSRFHHYHGSNIILFEENTVAYRKASFAHGLTFSEKPLLPGEIFLVEIEKTERGWSGHMRLGLTLLEPQAAALGDKGLPQYALPDLANNGRWENEPRVSVLGDGHVVKTPRGYISKMILKPQTVSQNGTPQGILPMDAGSRIGVMFVPCPKLNKNDQDLAEMHFIINGEDQGPCTKSIPYTRGLLHAVVDVYGTTKQVKIVQLYGVKSLQIICRDAILQNVKNGSVKSLPLPKSLKDFLLS
Summary
Uniprot
A0A2A4J0E5
A0A1E1WNJ0
A0A2H1VSD8
A0A0N1IP49
A0A194QCL0
A0A212EGZ6
+ More
A0A0L7LC47 A0A067RD59 S4P0Y9 A0A0K8TRB4 A0A2P8XWL0 A0A2J7PHX4 A0A1Q3FC50 Q16J25 B0WTA0 A0A023EMG2 A0A2M3ZIJ2 A0A2M4ALR8 A0A2M4BXN7 W5JX66 A0A0N7Z9L1 R4G441 A0A224XLX0 A0A069DYA3 A0A2M4BXP3 A0A084VUJ0 A0A0V0G9A6 A0A023FB48 A0A1B6K0D1 A0A182YFH5 T1DM69 A0A182LWP1 A0A182IS83 A0A182JUC0 A0A182NF95 A0A182Q5X8 A0A0A9W379 A0A2C9H8R4 A0A182UMF2 A0A182TKD4 Q7PW71 A0A1B6CWB2 A0A182KUS1 A0A1I8JUL2 A0A2C9GRV2 A0A182PDG2 A0A0C9RLE1 U5ETI4 A0A182WGW3 A0A1A9WI54 A0A088ANW8 A0A0A1WPE8 A0A2A3ERB8 A0A0K8VR84 A0A034WIJ8 A0A0M9ABD5 A0A1A9V6A5 A0A1B0GC56 A0A1A9X9M8 A0A1A9ZZH2 A0A0L8GPZ1 A0A1I8M6M8 E2C7N2 A0A182F0S0 A0A1S3IH06 A0A1I8Q4E8 A0A1S3IEV2 A0A1S3IFW6 A0A0L7QK48 A0A026WQP7 A0A158NB00 A0A310SP70 F4X5N2 A0A151K184 W8BH03 E9J976 A0A1Y1KDJ8 A0A3L8DMX3 A0A2R7WD65 A0A151IJ56 A0A154PNQ6 B4LN49 B4KTI3 V3ZIG5 A0A1W4XNH5 A0A170ZDI9 B4IH76 B3NQY1 B4QCJ4 B4PBU3 A0A1W4UZK3 Q9W112 B4J4E0 A0A0J7KPX5 A0A151I3D3 D6WTY4 B3MBK6 A0A151JMF3 A0A151WY32 A0A0M3QUL7
A0A0L7LC47 A0A067RD59 S4P0Y9 A0A0K8TRB4 A0A2P8XWL0 A0A2J7PHX4 A0A1Q3FC50 Q16J25 B0WTA0 A0A023EMG2 A0A2M3ZIJ2 A0A2M4ALR8 A0A2M4BXN7 W5JX66 A0A0N7Z9L1 R4G441 A0A224XLX0 A0A069DYA3 A0A2M4BXP3 A0A084VUJ0 A0A0V0G9A6 A0A023FB48 A0A1B6K0D1 A0A182YFH5 T1DM69 A0A182LWP1 A0A182IS83 A0A182JUC0 A0A182NF95 A0A182Q5X8 A0A0A9W379 A0A2C9H8R4 A0A182UMF2 A0A182TKD4 Q7PW71 A0A1B6CWB2 A0A182KUS1 A0A1I8JUL2 A0A2C9GRV2 A0A182PDG2 A0A0C9RLE1 U5ETI4 A0A182WGW3 A0A1A9WI54 A0A088ANW8 A0A0A1WPE8 A0A2A3ERB8 A0A0K8VR84 A0A034WIJ8 A0A0M9ABD5 A0A1A9V6A5 A0A1B0GC56 A0A1A9X9M8 A0A1A9ZZH2 A0A0L8GPZ1 A0A1I8M6M8 E2C7N2 A0A182F0S0 A0A1S3IH06 A0A1I8Q4E8 A0A1S3IEV2 A0A1S3IFW6 A0A0L7QK48 A0A026WQP7 A0A158NB00 A0A310SP70 F4X5N2 A0A151K184 W8BH03 E9J976 A0A1Y1KDJ8 A0A3L8DMX3 A0A2R7WD65 A0A151IJ56 A0A154PNQ6 B4LN49 B4KTI3 V3ZIG5 A0A1W4XNH5 A0A170ZDI9 B4IH76 B3NQY1 B4QCJ4 B4PBU3 A0A1W4UZK3 Q9W112 B4J4E0 A0A0J7KPX5 A0A151I3D3 D6WTY4 B3MBK6 A0A151JMF3 A0A151WY32 A0A0M3QUL7
Pubmed
26354079
22118469
26227816
24845553
23622113
26369729
+ More
29403074 17510324 24945155 20920257 23761445 27129103 26334808 24438588 25474469 25244985 25401762 26823975 12364791 14747013 17210077 20966253 25830018 25348373 25315136 20798317 24508170 21347285 21719571 24495485 21282665 28004739 30249741 17994087 23254933 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18362917 19820115
29403074 17510324 24945155 20920257 23761445 27129103 26334808 24438588 25474469 25244985 25401762 26823975 12364791 14747013 17210077 20966253 25830018 25348373 25315136 20798317 24508170 21347285 21719571 24495485 21282665 28004739 30249741 17994087 23254933 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18362917 19820115
EMBL
NWSH01004210
PCG65349.1
GDQN01009813
GDQN01002490
JAT81241.1
JAT88564.1
+ More
ODYU01004170 SOQ43755.1 KQ460772 KPJ12488.1 KQ459185 KPJ03278.1 AGBW02014995 OWR40758.1 JTDY01001758 KOB72965.1 KK852779 KDR16695.1 GAIX01009136 JAA83424.1 GDAI01000691 JAI16912.1 PYGN01001238 PSN36388.1 NEVH01025130 PNF15932.1 GFDL01009884 JAV25161.1 CH478035 EAT34272.1 DS232082 EDS34324.1 GAPW01003573 JAC10025.1 GGFM01007507 MBW28258.1 GGFK01008393 MBW41714.1 GGFJ01008642 MBW57783.1 ADMH02000010 ETN68100.1 GDKW01000243 JAI56352.1 GAHY01001675 JAA75835.1 GFTR01004342 JAW12084.1 GBGD01002540 JAC86349.1 GGFJ01008641 MBW57782.1 ATLV01016846 KE525120 KFB41634.1 GECL01001468 JAP04656.1 GBBI01000564 JAC18148.1 GECU01002801 JAT04906.1 GAMD01000056 JAB01535.1 AXCM01004220 AXCN02000824 GBHO01040707 GBHO01040702 GBRD01007172 GBRD01007171 GDHC01012540 JAG02897.1 JAG02902.1 JAG58649.1 JAQ06089.1 AAAB01008984 EAA14752.4 GEDC01019506 JAS17792.1 APCN01003737 GBYB01009100 JAG78867.1 GANO01002770 JAB57101.1 GBXI01013927 JAD00365.1 KZ288193 PBC34054.1 GDHF01010971 JAI41343.1 GAKP01004987 JAC53965.1 KQ435701 KOX80356.1 CCAG010018997 KQ420859 KOF79073.1 GL453395 EFN76015.1 KQ414986 KOC58998.1 KK107128 EZA58268.1 ADTU01010651 ADTU01010652 KQ762207 OAD56019.1 GL888730 EGI58237.1 KQ981225 KYN44369.1 GAMC01008533 JAB98022.1 GL769168 EFZ10640.1 GEZM01087615 JAV58578.1 QOIP01000006 RLU21573.1 KK854574 PTY17000.1 KQ977412 KYN02889.1 KQ435007 KZC13515.1 CH940648 EDW60053.1 CH933808 EDW08544.1 KB202284 ESO91073.1 GEMB01002332 JAS00851.1 CH480838 EDW49252.1 CH954179 EDV57064.1 CM000362 CM002911 EDX08610.1 KMY96422.1 CM000158 EDW92597.1 AE013599 AAF47265.3 CH916367 EDW01622.1 LBMM01004522 KMQ92326.1 KQ976506 KYM82839.1 KQ971352 EFA07350.1 CH902619 EDV37137.1 KQ978966 KYN26905.1 KQ982665 KYQ52601.1 CP012524 ALC40889.1
ODYU01004170 SOQ43755.1 KQ460772 KPJ12488.1 KQ459185 KPJ03278.1 AGBW02014995 OWR40758.1 JTDY01001758 KOB72965.1 KK852779 KDR16695.1 GAIX01009136 JAA83424.1 GDAI01000691 JAI16912.1 PYGN01001238 PSN36388.1 NEVH01025130 PNF15932.1 GFDL01009884 JAV25161.1 CH478035 EAT34272.1 DS232082 EDS34324.1 GAPW01003573 JAC10025.1 GGFM01007507 MBW28258.1 GGFK01008393 MBW41714.1 GGFJ01008642 MBW57783.1 ADMH02000010 ETN68100.1 GDKW01000243 JAI56352.1 GAHY01001675 JAA75835.1 GFTR01004342 JAW12084.1 GBGD01002540 JAC86349.1 GGFJ01008641 MBW57782.1 ATLV01016846 KE525120 KFB41634.1 GECL01001468 JAP04656.1 GBBI01000564 JAC18148.1 GECU01002801 JAT04906.1 GAMD01000056 JAB01535.1 AXCM01004220 AXCN02000824 GBHO01040707 GBHO01040702 GBRD01007172 GBRD01007171 GDHC01012540 JAG02897.1 JAG02902.1 JAG58649.1 JAQ06089.1 AAAB01008984 EAA14752.4 GEDC01019506 JAS17792.1 APCN01003737 GBYB01009100 JAG78867.1 GANO01002770 JAB57101.1 GBXI01013927 JAD00365.1 KZ288193 PBC34054.1 GDHF01010971 JAI41343.1 GAKP01004987 JAC53965.1 KQ435701 KOX80356.1 CCAG010018997 KQ420859 KOF79073.1 GL453395 EFN76015.1 KQ414986 KOC58998.1 KK107128 EZA58268.1 ADTU01010651 ADTU01010652 KQ762207 OAD56019.1 GL888730 EGI58237.1 KQ981225 KYN44369.1 GAMC01008533 JAB98022.1 GL769168 EFZ10640.1 GEZM01087615 JAV58578.1 QOIP01000006 RLU21573.1 KK854574 PTY17000.1 KQ977412 KYN02889.1 KQ435007 KZC13515.1 CH940648 EDW60053.1 CH933808 EDW08544.1 KB202284 ESO91073.1 GEMB01002332 JAS00851.1 CH480838 EDW49252.1 CH954179 EDV57064.1 CM000362 CM002911 EDX08610.1 KMY96422.1 CM000158 EDW92597.1 AE013599 AAF47265.3 CH916367 EDW01622.1 LBMM01004522 KMQ92326.1 KQ976506 KYM82839.1 KQ971352 EFA07350.1 CH902619 EDV37137.1 KQ978966 KYN26905.1 KQ982665 KYQ52601.1 CP012524 ALC40889.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000027135
+ More
UP000245037 UP000235965 UP000008820 UP000002320 UP000000673 UP000030765 UP000076408 UP000075883 UP000075880 UP000075881 UP000075884 UP000075886 UP000076407 UP000075903 UP000075902 UP000007062 UP000075882 UP000075900 UP000075840 UP000075885 UP000075920 UP000091820 UP000005203 UP000242457 UP000053105 UP000078200 UP000092444 UP000092443 UP000092445 UP000053454 UP000095301 UP000008237 UP000069272 UP000085678 UP000095300 UP000053825 UP000053097 UP000005205 UP000007755 UP000078541 UP000279307 UP000078542 UP000076502 UP000008792 UP000009192 UP000030746 UP000192223 UP000001292 UP000008711 UP000000304 UP000002282 UP000192221 UP000000803 UP000001070 UP000036403 UP000078540 UP000007266 UP000007801 UP000078492 UP000075809 UP000092553
UP000245037 UP000235965 UP000008820 UP000002320 UP000000673 UP000030765 UP000076408 UP000075883 UP000075880 UP000075881 UP000075884 UP000075886 UP000076407 UP000075903 UP000075902 UP000007062 UP000075882 UP000075900 UP000075840 UP000075885 UP000075920 UP000091820 UP000005203 UP000242457 UP000053105 UP000078200 UP000092444 UP000092443 UP000092445 UP000053454 UP000095301 UP000008237 UP000069272 UP000085678 UP000095300 UP000053825 UP000053097 UP000005205 UP000007755 UP000078541 UP000279307 UP000078542 UP000076502 UP000008792 UP000009192 UP000030746 UP000192223 UP000001292 UP000008711 UP000000304 UP000002282 UP000192221 UP000000803 UP000001070 UP000036403 UP000078540 UP000007266 UP000007801 UP000078492 UP000075809 UP000092553
PRIDE
Interpro
ProteinModelPortal
A0A2A4J0E5
A0A1E1WNJ0
A0A2H1VSD8
A0A0N1IP49
A0A194QCL0
A0A212EGZ6
+ More
A0A0L7LC47 A0A067RD59 S4P0Y9 A0A0K8TRB4 A0A2P8XWL0 A0A2J7PHX4 A0A1Q3FC50 Q16J25 B0WTA0 A0A023EMG2 A0A2M3ZIJ2 A0A2M4ALR8 A0A2M4BXN7 W5JX66 A0A0N7Z9L1 R4G441 A0A224XLX0 A0A069DYA3 A0A2M4BXP3 A0A084VUJ0 A0A0V0G9A6 A0A023FB48 A0A1B6K0D1 A0A182YFH5 T1DM69 A0A182LWP1 A0A182IS83 A0A182JUC0 A0A182NF95 A0A182Q5X8 A0A0A9W379 A0A2C9H8R4 A0A182UMF2 A0A182TKD4 Q7PW71 A0A1B6CWB2 A0A182KUS1 A0A1I8JUL2 A0A2C9GRV2 A0A182PDG2 A0A0C9RLE1 U5ETI4 A0A182WGW3 A0A1A9WI54 A0A088ANW8 A0A0A1WPE8 A0A2A3ERB8 A0A0K8VR84 A0A034WIJ8 A0A0M9ABD5 A0A1A9V6A5 A0A1B0GC56 A0A1A9X9M8 A0A1A9ZZH2 A0A0L8GPZ1 A0A1I8M6M8 E2C7N2 A0A182F0S0 A0A1S3IH06 A0A1I8Q4E8 A0A1S3IEV2 A0A1S3IFW6 A0A0L7QK48 A0A026WQP7 A0A158NB00 A0A310SP70 F4X5N2 A0A151K184 W8BH03 E9J976 A0A1Y1KDJ8 A0A3L8DMX3 A0A2R7WD65 A0A151IJ56 A0A154PNQ6 B4LN49 B4KTI3 V3ZIG5 A0A1W4XNH5 A0A170ZDI9 B4IH76 B3NQY1 B4QCJ4 B4PBU3 A0A1W4UZK3 Q9W112 B4J4E0 A0A0J7KPX5 A0A151I3D3 D6WTY4 B3MBK6 A0A151JMF3 A0A151WY32 A0A0M3QUL7
A0A0L7LC47 A0A067RD59 S4P0Y9 A0A0K8TRB4 A0A2P8XWL0 A0A2J7PHX4 A0A1Q3FC50 Q16J25 B0WTA0 A0A023EMG2 A0A2M3ZIJ2 A0A2M4ALR8 A0A2M4BXN7 W5JX66 A0A0N7Z9L1 R4G441 A0A224XLX0 A0A069DYA3 A0A2M4BXP3 A0A084VUJ0 A0A0V0G9A6 A0A023FB48 A0A1B6K0D1 A0A182YFH5 T1DM69 A0A182LWP1 A0A182IS83 A0A182JUC0 A0A182NF95 A0A182Q5X8 A0A0A9W379 A0A2C9H8R4 A0A182UMF2 A0A182TKD4 Q7PW71 A0A1B6CWB2 A0A182KUS1 A0A1I8JUL2 A0A2C9GRV2 A0A182PDG2 A0A0C9RLE1 U5ETI4 A0A182WGW3 A0A1A9WI54 A0A088ANW8 A0A0A1WPE8 A0A2A3ERB8 A0A0K8VR84 A0A034WIJ8 A0A0M9ABD5 A0A1A9V6A5 A0A1B0GC56 A0A1A9X9M8 A0A1A9ZZH2 A0A0L8GPZ1 A0A1I8M6M8 E2C7N2 A0A182F0S0 A0A1S3IH06 A0A1I8Q4E8 A0A1S3IEV2 A0A1S3IFW6 A0A0L7QK48 A0A026WQP7 A0A158NB00 A0A310SP70 F4X5N2 A0A151K184 W8BH03 E9J976 A0A1Y1KDJ8 A0A3L8DMX3 A0A2R7WD65 A0A151IJ56 A0A154PNQ6 B4LN49 B4KTI3 V3ZIG5 A0A1W4XNH5 A0A170ZDI9 B4IH76 B3NQY1 B4QCJ4 B4PBU3 A0A1W4UZK3 Q9W112 B4J4E0 A0A0J7KPX5 A0A151I3D3 D6WTY4 B3MBK6 A0A151JMF3 A0A151WY32 A0A0M3QUL7
PDB
4KG0
E-value=9.45473e-10,
Score=149
Ontologies
PANTHER
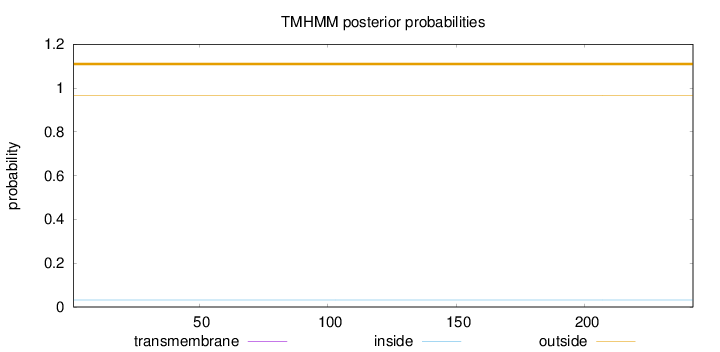
Topology
Length:
242
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00152
Exp number, first 60 AAs:
0.000830000000000001
Total prob of N-in:
0.03251
outside
1 - 242
Population Genetic Test Statistics
Pi
140.382065
Theta
183.70847
Tajima's D
-0.572949
CLR
342.045784
CSRT
0.224588770561472
Interpretation
Uncertain
