Pre Gene Modal
BGIBMGA004108
Annotation
PREDICTED:_RNA_polymerase_II_subunit_A_C-terminal_domain_phosphatase_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.654
Sequence
CDS
ATGGCCGACAAAACTATACCTATTTTTGTTCCATCAGAAAAGCCTGTGAAACTCGCAAAGTGGAAAATAAAACAAGGTACTGTGGTAACACAAGGCCAAGTATTATTTTTATACAAAGATTTAGCGGGCGATGGTGAAGAGTTAAAGAAATTTAAATCATCACACGCTGGTACTGTGTCATCGATCAAGGTGAAAGAAGGAGACGTAGTAGAACCTAGTTCGGCTGTAGCAGATTTGGAAGAATGCCGCCATCCAACAGTGATGATGGAGATGTGCGCGGAGTGTGGAGCAGACCTCCGCAGCGAGGAGACCAAGAAGCTGGACGTGGCCATTGTACCAATGGTACACTCTGTACCTGAATTAAAAGTGTCTGAAGAACTGGCACAAAAACTAGGTAAAGAGGATGCTGAGCGTCTTCTGAAAGACAGGAAATTAGTACTTCTGGTAGATTTAGATCAAACATTAGTTCACACTACTAATGACAATATACCACCCAATCTAAAGGGAGTATTACACTTCTTCCTAAGAGGACCTGGAGGGCCAGGGCGTTGGTGTCATACGAGGCTTCGACCACGCACTCAAGAGTTTCTTGAATCAGCAGCAAAGAACTATGAACTTCACATATGTACATTTGGAGCCAGGCAATATGCACATGCCGTAGCTGAGCTCCTAGATCCGGAAAAGAAATATTTCTCACATAGGATACTATCCAGAGATGAATGCTTTGATCCTAGAACTAAAGCAGCCAATCTGAAAGCACTGTTTCCTTGTGGTGACAACATGGTTTGCATTATTGACGATCGGGAAGATGTGTGGAGACATGCTGCTAATCTAATTCATGTCAGGCCTTACTCATTTTTTCAATCGACCGGCGATATCAATGCACCGCCACCAATTGAAGAGAAACCAAAATTGACGATCGGCAAAAATGGTACTCAACCACAAGTACAAGAAATGCCGACTTTAGATGCAGAACCAGATGAAAATCTAGAAAAACCTGCTAACAACTTTGAAAAACCTACTGAAGGTAGCAACGGAAAAGTCATAAAAGAAGTAATGGAAGAGAAGCCAGTTGATATTAAGGAAGGCAATGAAAAAATTGAGGAAGTTGAATTACCGCCAGCTGAGCCAGTATGGACAGAGACATCTGATGGACAGATAGAGGTGGAAGATCCTGATGATTATCTTATCTATTTGGAGGATATTTTGAAGAGGATACATAAGCATTTTTATGATGTCTATGACAAAATGGGTCAGAAACAGATACCAGACCTTAAAATAGTAATACCTGAAGTTAAAAGTGAAGTGTTGACTGGAACTAGTTTAGTGTTTAGTGGTTTAGTGCCTACACATCAGAAACTGGAAACTTCTAGAGCCTATCTTGTGGCGAGAAGTCTTGGTGCTGAGGTGACACAGGATTTTACTGATAAAACAACTCATTTAGTAGCTGTTAGAGCAGGCACGGCAAAAGTAAACACAAGTAGGCGAATGACTGAGAAGAAATCCAGCAAATTACATGTTGTTACACCAGATTGGCTGTGGTCATGTGCGGAACGATGGGAACGAGTGGAAGAGAAATTATATACATTACAAAGAGGTGGACAGAGCAGCTGTCGTCGGCCGCCGGCGCACTGCAGCAGCCCGCCCCCGCCGCCCGCGCCCGTCGCCGCCCTCCGCCGCCGCACGCCCTCGGGACGCTTCATGGACACCATCAACCCGCTGCTCTCCTTCTCCAGCGACGACATTGCTGATATGGACCGAGAGGTTGAAGATATTTTCAATGAATCAGATGAGAGTTCCACTGATGATGAGAAACCATCCCAAGATGATGTTGATGATGAAGAAAACTTAAAGGAAGATCAACTAATTAGGATAGATGAAGAAAGTACTCAAGACAGATTACAAGAAACGCAAGATAGCTCTAATGAATCAGAAAACGACGAAGAACAGCCAAGACCACGGAAACGACCGCGCAAATCGACACCGCCGTCAGACGAAGAGCCGCCGGATGATGACGACAGCTCTTGGAACTTGATGGGCGCGGCACTTGAAAGAGAGTTCCTCGCGCAAGACTGA
Protein
MADKTIPIFVPSEKPVKLAKWKIKQGTVVTQGQVLFLYKDLAGDGEELKKFKSSHAGTVSSIKVKEGDVVEPSSAVADLEECRHPTVMMEMCAECGADLRSEETKKLDVAIVPMVHSVPELKVSEELAQKLGKEDAERLLKDRKLVLLVDLDQTLVHTTNDNIPPNLKGVLHFFLRGPGGPGRWCHTRLRPRTQEFLESAAKNYELHICTFGARQYAHAVAELLDPEKKYFSHRILSRDECFDPRTKAANLKALFPCGDNMVCIIDDREDVWRHAANLIHVRPYSFFQSTGDINAPPPIEEKPKLTIGKNGTQPQVQEMPTLDAEPDENLEKPANNFEKPTEGSNGKVIKEVMEEKPVDIKEGNEKIEEVELPPAEPVWTETSDGQIEVEDPDDYLIYLEDILKRIHKHFYDVYDKMGQKQIPDLKIVIPEVKSEVLTGTSLVFSGLVPTHQKLETSRAYLVARSLGAEVTQDFTDKTTHLVAVRAGTAKVNTSRRMTEKKSSKLHVVTPDWLWSCAERWERVEEKLYTLQRGGQSSCRRPPAHCSSPPPPPAPVAALRRRTPSGRFMDTINPLLSFSSDDIADMDREVEDIFNESDESSTDDEKPSQDDVDDEENLKEDQLIRIDEESTQDRLQETQDSSNESENDEEQPRPRKRPRKSTPPSDEEPPDDDDSSWNLMGAALEREFLAQD
Summary
Uniprot
H9J3M1
A0A2A4JIK0
A0A2H1VB77
A0A194QEB2
A0A212EGZ8
A0A1Y1NDS0
+ More
D6WSR3 A0A1A9WPA2 A0A1B0FN26 A0A1A9YJ08 A0A1B0B694 A0A1A9UV41 A0A1B0AAP6 A0A182H2D7 A0A0M4E463 A0A1Q3EZ87 B4MP75 B0WCB3 A0A310SRQ9 A0A088AJB9 Q16EJ3 A0A0L7R7V8 E2B8Q6 A0A336LUZ3 A0A195CLI4 A0A2A3E837 A0A1S4G3I0 E9J520 F4W821 E2AXU1 E9H914 A0A151X4I5 A0A195EYG3 J9JN04 A0A2H8THJ5 A0A2S2R246 A0A158NDS2 A0A2S2R779 A0A195BR06 A0A1J1IT20 A0A2H8TU57 B4N3A4 A0A0K2UNY2 A0A0M8ZTC9 A0A0J7KSU8 A0A226F5J3 A0A3S0ZVS2 T1IZ83 V3ZU10 A0A0P5PDP6 A0A0P4YXM0 A0A0P5PBA0 A0A0P5QR90 A0A0P6D6D1 A0A0P5VFB5 A0A162T9U8 A0A0P6D6E1 A0A0P6BHT7 A0A0P5S3I0 A0A0P5KAY0 A0A0P6H9T5 A0A0P5SIF4 A0A1S3H005 A0A1S3K5N8 A0A0K8TI11 A0A0A9YEQ7 A0A146LM22 A0A0V0GBR8 A0A3S3P404 T1K4P5 A0A0N8BLQ5 A0A0P5LRN8 A0A0P5YXX2 A0A224XMS6 A0A0P5W7W5 T1ESM4 A0A1W0X3B0 A0A132AH82 A0A1D1VE07
D6WSR3 A0A1A9WPA2 A0A1B0FN26 A0A1A9YJ08 A0A1B0B694 A0A1A9UV41 A0A1B0AAP6 A0A182H2D7 A0A0M4E463 A0A1Q3EZ87 B4MP75 B0WCB3 A0A310SRQ9 A0A088AJB9 Q16EJ3 A0A0L7R7V8 E2B8Q6 A0A336LUZ3 A0A195CLI4 A0A2A3E837 A0A1S4G3I0 E9J520 F4W821 E2AXU1 E9H914 A0A151X4I5 A0A195EYG3 J9JN04 A0A2H8THJ5 A0A2S2R246 A0A158NDS2 A0A2S2R779 A0A195BR06 A0A1J1IT20 A0A2H8TU57 B4N3A4 A0A0K2UNY2 A0A0M8ZTC9 A0A0J7KSU8 A0A226F5J3 A0A3S0ZVS2 T1IZ83 V3ZU10 A0A0P5PDP6 A0A0P4YXM0 A0A0P5PBA0 A0A0P5QR90 A0A0P6D6D1 A0A0P5VFB5 A0A162T9U8 A0A0P6D6E1 A0A0P6BHT7 A0A0P5S3I0 A0A0P5KAY0 A0A0P6H9T5 A0A0P5SIF4 A0A1S3H005 A0A1S3K5N8 A0A0K8TI11 A0A0A9YEQ7 A0A146LM22 A0A0V0GBR8 A0A3S3P404 T1K4P5 A0A0N8BLQ5 A0A0P5LRN8 A0A0P5YXX2 A0A224XMS6 A0A0P5W7W5 T1ESM4 A0A1W0X3B0 A0A132AH82 A0A1D1VE07
Pubmed
EMBL
BABH01021627
NWSH01001346
PCG71596.1
ODYU01001611
SOQ38077.1
KQ459185
+ More
KPJ03285.1 AGBW02014995 OWR40763.1 GEZM01005547 JAV96104.1 KQ971354 EFA07148.1 CCAG010002874 JXJN01009035 JXUM01105103 JXUM01105104 KQ564939 KXJ71534.1 CP012524 ALC40754.1 GFDL01014428 JAV20617.1 CH963848 EDW73914.1 DS231886 EDS43358.1 KQ760629 OAD59738.1 CH478692 EAT32653.1 KQ414637 KOC66965.1 GL446361 EFN87962.1 UFQS01000211 UFQT01000211 SSX01374.1 SSX21754.1 KQ977600 KYN01568.1 KZ288335 PBC27860.1 GL768123 EFZ12041.1 GL887888 EGI69691.1 GL443699 EFN61749.1 GL732607 EFX71774.1 KQ982548 KYQ55199.1 KQ981906 KYN33258.1 ABLF02035887 GFXV01001788 MBW13593.1 GGMS01014875 MBY84078.1 ADTU01012852 GGMS01016652 MBY85855.1 KQ976424 KYM88394.1 CVRI01000059 CRL03381.1 GFXV01005980 MBW17785.1 CH964062 EDW78843.2 HACA01022241 CDW39602.1 KQ435851 KOX70730.1 LBMM01003490 KMQ93472.1 LNIX01000001 OXA65075.1 RQTK01000215 RUS84178.1 AFFK01020413 KB202823 ESO87837.1 GDIQ01132285 JAL19441.1 GDIP01225668 JAI97733.1 GDIQ01137017 JAL14709.1 GDIQ01130978 JAL20748.1 GDIQ01082137 JAN12600.1 GDIP01115821 JAL87893.1 LRGB01000005 KZS22024.1 GDIQ01082136 JAN12601.1 GDIP01028946 JAM74769.1 GDIQ01092886 JAL58840.1 GDIQ01193151 JAK58574.1 GDIQ01022616 JAN72121.1 GDIQ01092887 JAL58839.1 GBRD01000574 JAG65247.1 GBHO01013503 JAG30101.1 GDHC01010763 JAQ07866.1 GECL01000593 JAP05531.1 NCKU01001628 RWS11626.1 CAEY01001579 GDIQ01165419 JAK86306.1 GDIQ01166959 JAK84766.1 GDIP01053603 JAM50112.1 GFTR01007127 JAW09299.1 GDIP01090134 JAM13581.1 AMQM01001081 KB097143 ESN98945.1 MTYJ01000020 OQV21995.1 JXLN01014887 KPM10287.1 BDGG01000005 GAU99025.1
KPJ03285.1 AGBW02014995 OWR40763.1 GEZM01005547 JAV96104.1 KQ971354 EFA07148.1 CCAG010002874 JXJN01009035 JXUM01105103 JXUM01105104 KQ564939 KXJ71534.1 CP012524 ALC40754.1 GFDL01014428 JAV20617.1 CH963848 EDW73914.1 DS231886 EDS43358.1 KQ760629 OAD59738.1 CH478692 EAT32653.1 KQ414637 KOC66965.1 GL446361 EFN87962.1 UFQS01000211 UFQT01000211 SSX01374.1 SSX21754.1 KQ977600 KYN01568.1 KZ288335 PBC27860.1 GL768123 EFZ12041.1 GL887888 EGI69691.1 GL443699 EFN61749.1 GL732607 EFX71774.1 KQ982548 KYQ55199.1 KQ981906 KYN33258.1 ABLF02035887 GFXV01001788 MBW13593.1 GGMS01014875 MBY84078.1 ADTU01012852 GGMS01016652 MBY85855.1 KQ976424 KYM88394.1 CVRI01000059 CRL03381.1 GFXV01005980 MBW17785.1 CH964062 EDW78843.2 HACA01022241 CDW39602.1 KQ435851 KOX70730.1 LBMM01003490 KMQ93472.1 LNIX01000001 OXA65075.1 RQTK01000215 RUS84178.1 AFFK01020413 KB202823 ESO87837.1 GDIQ01132285 JAL19441.1 GDIP01225668 JAI97733.1 GDIQ01137017 JAL14709.1 GDIQ01130978 JAL20748.1 GDIQ01082137 JAN12600.1 GDIP01115821 JAL87893.1 LRGB01000005 KZS22024.1 GDIQ01082136 JAN12601.1 GDIP01028946 JAM74769.1 GDIQ01092886 JAL58840.1 GDIQ01193151 JAK58574.1 GDIQ01022616 JAN72121.1 GDIQ01092887 JAL58839.1 GBRD01000574 JAG65247.1 GBHO01013503 JAG30101.1 GDHC01010763 JAQ07866.1 GECL01000593 JAP05531.1 NCKU01001628 RWS11626.1 CAEY01001579 GDIQ01165419 JAK86306.1 GDIQ01166959 JAK84766.1 GDIP01053603 JAM50112.1 GFTR01007127 JAW09299.1 GDIP01090134 JAM13581.1 AMQM01001081 KB097143 ESN98945.1 MTYJ01000020 OQV21995.1 JXLN01014887 KPM10287.1 BDGG01000005 GAU99025.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000007266
UP000091820
+ More
UP000092444 UP000092443 UP000092460 UP000078200 UP000092445 UP000069940 UP000249989 UP000092553 UP000007798 UP000002320 UP000005203 UP000008820 UP000053825 UP000008237 UP000078542 UP000242457 UP000007755 UP000000311 UP000000305 UP000075809 UP000078541 UP000007819 UP000005205 UP000078540 UP000183832 UP000053105 UP000036403 UP000198287 UP000271974 UP000030746 UP000076858 UP000085678 UP000285301 UP000015104 UP000015101 UP000186922
UP000092444 UP000092443 UP000092460 UP000078200 UP000092445 UP000069940 UP000249989 UP000092553 UP000007798 UP000002320 UP000005203 UP000008820 UP000053825 UP000008237 UP000078542 UP000242457 UP000007755 UP000000311 UP000000305 UP000075809 UP000078541 UP000007819 UP000005205 UP000078540 UP000183832 UP000053105 UP000036403 UP000198287 UP000271974 UP000030746 UP000076858 UP000085678 UP000285301 UP000015104 UP000015101 UP000186922
PRIDE
Pfam
Interpro
IPR011947
FCP1_euk
+ More
IPR011053 Single_hybrid_motif
IPR004274 FCP1_dom
IPR036412 HAD-like_sf
IPR036420 BRCT_dom_sf
IPR001357 BRCT_dom
IPR023214 HAD_sf
IPR015388 FCP1_C
IPR039189 Fcp1
IPR039562 MFP_biotin_lipoyl_2
IPR000089 Biotin_lipoyl
IPR024969 Rpn11/EIF3F_C
IPR033858 MPN_RPN7_8
IPR000555 JAMM/MPN+_dom
IPR037518 MPN
IPR011053 Single_hybrid_motif
IPR004274 FCP1_dom
IPR036412 HAD-like_sf
IPR036420 BRCT_dom_sf
IPR001357 BRCT_dom
IPR023214 HAD_sf
IPR015388 FCP1_C
IPR039189 Fcp1
IPR039562 MFP_biotin_lipoyl_2
IPR000089 Biotin_lipoyl
IPR024969 Rpn11/EIF3F_C
IPR033858 MPN_RPN7_8
IPR000555 JAMM/MPN+_dom
IPR037518 MPN
Gene 3D
ProteinModelPortal
H9J3M1
A0A2A4JIK0
A0A2H1VB77
A0A194QEB2
A0A212EGZ8
A0A1Y1NDS0
+ More
D6WSR3 A0A1A9WPA2 A0A1B0FN26 A0A1A9YJ08 A0A1B0B694 A0A1A9UV41 A0A1B0AAP6 A0A182H2D7 A0A0M4E463 A0A1Q3EZ87 B4MP75 B0WCB3 A0A310SRQ9 A0A088AJB9 Q16EJ3 A0A0L7R7V8 E2B8Q6 A0A336LUZ3 A0A195CLI4 A0A2A3E837 A0A1S4G3I0 E9J520 F4W821 E2AXU1 E9H914 A0A151X4I5 A0A195EYG3 J9JN04 A0A2H8THJ5 A0A2S2R246 A0A158NDS2 A0A2S2R779 A0A195BR06 A0A1J1IT20 A0A2H8TU57 B4N3A4 A0A0K2UNY2 A0A0M8ZTC9 A0A0J7KSU8 A0A226F5J3 A0A3S0ZVS2 T1IZ83 V3ZU10 A0A0P5PDP6 A0A0P4YXM0 A0A0P5PBA0 A0A0P5QR90 A0A0P6D6D1 A0A0P5VFB5 A0A162T9U8 A0A0P6D6E1 A0A0P6BHT7 A0A0P5S3I0 A0A0P5KAY0 A0A0P6H9T5 A0A0P5SIF4 A0A1S3H005 A0A1S3K5N8 A0A0K8TI11 A0A0A9YEQ7 A0A146LM22 A0A0V0GBR8 A0A3S3P404 T1K4P5 A0A0N8BLQ5 A0A0P5LRN8 A0A0P5YXX2 A0A224XMS6 A0A0P5W7W5 T1ESM4 A0A1W0X3B0 A0A132AH82 A0A1D1VE07
D6WSR3 A0A1A9WPA2 A0A1B0FN26 A0A1A9YJ08 A0A1B0B694 A0A1A9UV41 A0A1B0AAP6 A0A182H2D7 A0A0M4E463 A0A1Q3EZ87 B4MP75 B0WCB3 A0A310SRQ9 A0A088AJB9 Q16EJ3 A0A0L7R7V8 E2B8Q6 A0A336LUZ3 A0A195CLI4 A0A2A3E837 A0A1S4G3I0 E9J520 F4W821 E2AXU1 E9H914 A0A151X4I5 A0A195EYG3 J9JN04 A0A2H8THJ5 A0A2S2R246 A0A158NDS2 A0A2S2R779 A0A195BR06 A0A1J1IT20 A0A2H8TU57 B4N3A4 A0A0K2UNY2 A0A0M8ZTC9 A0A0J7KSU8 A0A226F5J3 A0A3S0ZVS2 T1IZ83 V3ZU10 A0A0P5PDP6 A0A0P4YXM0 A0A0P5PBA0 A0A0P5QR90 A0A0P6D6D1 A0A0P5VFB5 A0A162T9U8 A0A0P6D6E1 A0A0P6BHT7 A0A0P5S3I0 A0A0P5KAY0 A0A0P6H9T5 A0A0P5SIF4 A0A1S3H005 A0A1S3K5N8 A0A0K8TI11 A0A0A9YEQ7 A0A146LM22 A0A0V0GBR8 A0A3S3P404 T1K4P5 A0A0N8BLQ5 A0A0P5LRN8 A0A0P5YXX2 A0A224XMS6 A0A0P5W7W5 T1ESM4 A0A1W0X3B0 A0A132AH82 A0A1D1VE07
PDB
3EF1
E-value=1.13206e-39,
Score=413
Ontologies
GO
PANTHER
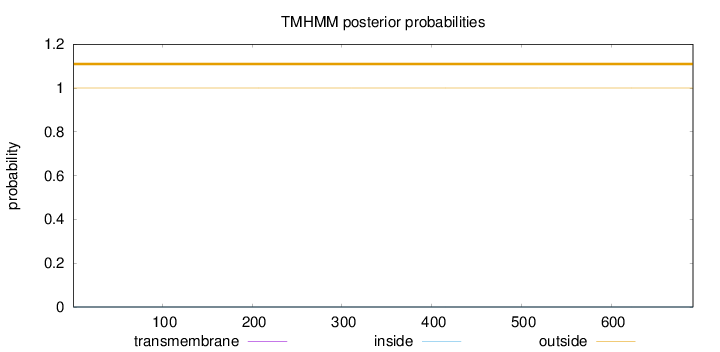
Topology
Length:
691
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00038
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00015
outside
1 - 691
Population Genetic Test Statistics
Pi
218.642066
Theta
186.300498
Tajima's D
0.425237
CLR
0.191309
CSRT
0.485075746212689
Interpretation
Uncertain
