Gene
KWMTBOMO11605
Pre Gene Modal
BGIBMGA004014
Annotation
PREDICTED:_collagen_type_IV_alpha-3-binding_protein_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.879 Nuclear Reliability : 2.396
Sequence
CDS
ATGATTGACGAAAACATCACAGTCAGCGATAGTTCTGATTTAGAAGAGGGCGGAACTCCTGAACTTCGCGGATACCTAAGCAAATGGACAAACTATATACACGGCTGGCAGGATAGATTTATTGTCCTTAAGGATTCTACTTTATCTTATTACAAGAATGAACTCGAAAGCAATTTAGGATGCCGCGGTGCCTTATGTCTGAAAAAAGCTAAAGTCAAGCCTCACGAGTTTGATGACTGTAGATTCGATGTTTCTGTTAGCGACTGCGTTTGGTACTTAAGAGCGTCTAATCCAGAGGAGAAACAGCAATGGATTGATATTCTAGAGTCTTTCAAAGTTGAATCAGGGTACGGTACAAGTTCAGATAGTAGCTCAAATGGAGGAGGTCTTCAAAGGCATGGTTCTGCCACTTCACTACAGTCTACAGGGTCACACGGTCACTCTACACGTCGCCTCACAGAGAAAATCACTGAGCTGGAAACGTACAATGAGCTTCTGTCAAAGCAAGCAATACAACTGCAACAATACTTTGATATGTGTATCGATTCTTACCCAAAAATTGATGATGCTAATGATGCGATAGATGCTGTTAAATTAGCCGATGGCAACGATCTCAGAGTGCACGCAGGCGAAGTGCGGGCGACGAGTACGTCGTTCCGAGCGACGTGCGGCGCGACGGTGGCGACGCTGCAGCACTGCGTCGAACTGTTGCGTCGACGCGAGAAGCAGGCGAGACAAGCCGAGGACTTATATATGGCTCTCAAGACTCAACTAAATGAAGCGAGATTAGCATCCAAACCCGGACCAGACCACGAGGAGGGTCCACACTCCACCCTCCCCGACGACGAGTTCCACGATGCGATCGAGACAGGCTTCGACAAAATGGAGGAGGAGCGGTGCACAAGGGTGGTGCCGACCGTACACACCCGCGACCAGATTGAATTGCCACCGCCGGCCATCGATAACCGTCTCACCGTACATCCCCTGTGGCCGGAGATTGACAGGATATCGACAGAACAAATACAGGCCGCGTTCGAAGGTGTCGGCGGTGAAATCGGGTGGCAGTTGTTTGCTGAAGAGGGGGACATGAGGATGTACAGAAGGGAAGTGGAAGTCGACGGCATGGTGATGGATCCTCTGAAGGCGATGCACAAGGTGCGTGGCGTCTCTGCGCGTGAAATGTGTCATTACTTCTTCAACCCGAGATACAGATATGAATGGGAAACTACCCTAGAACAGATGACGATAGTAGAAGAGATCTCATCAGATGCGCTGGTGTTCCACCAAACGTTCAAGCGGATCTGGCCGGCGTCGCAGCGGGACGCACTGTTCTGGTCACACGTGAGGGCCTCCAGCGAAAACACCTTCGCTGTAACCAACCATTCCACTACCAACACGGAATACCCAGCGAACTCTAGTGCGTGCATCCGTTTGTTCGTGACCGTGTGTCTGGCTTGCCGCAGCACGTACCCGGCCGGCGAGCAGCCCACGCGGGATAATATCACCACGAGCATCGCCTATTGCAGTACAGTGAACCCTGGAGGTTGGGCACCGGCCGGCGTGCTCCGTGCCGTGTACAAACGTGAATATCCCAAGTTCCTGAAGCGTTTCACCGGCTACGTCGTCGACCAGTGCAAGGACAAACCTCTCGCCATCTAG
Protein
MIDENITVSDSSDLEEGGTPELRGYLSKWTNYIHGWQDRFIVLKDSTLSYYKNELESNLGCRGALCLKKAKVKPHEFDDCRFDVSVSDCVWYLRASNPEEKQQWIDILESFKVESGYGTSSDSSSNGGGLQRHGSATSLQSTGSHGHSTRRLTEKITELETYNELLSKQAIQLQQYFDMCIDSYPKIDDANDAIDAVKLADGNDLRVHAGEVRATSTSFRATCGATVATLQHCVELLRRREKQARQAEDLYMALKTQLNEARLASKPGPDHEEGPHSTLPDDEFHDAIETGFDKMEEERCTRVVPTVHTRDQIELPPPAIDNRLTVHPLWPEIDRISTEQIQAAFEGVGGEIGWQLFAEEGDMRMYRREVEVDGMVMDPLKAMHKVRGVSAREMCHYFFNPRYRYEWETTLEQMTIVEEISSDALVFHQTFKRIWPASQRDALFWSHVRASSENTFAVTNHSTTNTEYPANSSACIRLFVTVCLACRSTYPAGEQPTRDNITTSIAYCSTVNPGGWAPAGVLRAVYKREYPKFLKRFTGYVVDQCKDKPLAI
Summary
Uniprot
A0A194QIK7
A0A0N1IDF5
A0A2A4JHR7
A0A2M4BII1
A0A182U1V8
A0A182KQA6
+ More
A0A2C9GPI7 A0A182VBG3 A0A2M3Z7E0 Q5TWP3 A0A023EVC7 A0A2M4AJZ7 A0A2M4AJR3 A0A1L8DMF9 A0A2M4CUU7 A0A2M4CUE5 A0A182YEH8 A0A1L8DMA7 A0A182QKK5 A0A182JB80 A0A2P8YYJ8 D6WR07 A0A151HYC6 E2BKT8 E2A0T0 A0A195ES80 A0A154PND9 A0A0L7RBJ3 A0A026WKM2 A0A151X559 A0A151IC13 F4X4T6 E9J4P7 A0A1Y1MER9 A0A088AJ90 A0A2A3ETZ5 A0A310S662 A0A1B6ECM8 A0A0A9YTN1 A0A0K8SCK0 A0A1B6EKV6 A0A1B6E8C9 J3JY04 A0A1W4WM14 Q9Y128 B4PG56 B3NFV1 A0A0K8U4A9 B4HJF4 A0A0J9RR46 A0A0A1WWK4 A0A034WQ32 B4L944 A0A1W4USF6 A0A0L0CAD6 B4LCA3 B4J205 B3M9P2 A0A1J1HZD1 A0A3B0KLS5 W8BMR1 Q29F82 A0A336MKJ2 A0A1A9UCS9 A0A1A9Z820 A0A0M4EKZ1 B4MKS1 E9FT92 A0A1A9Y4S9 A0A1B0BE36 E0VRU4 A0A1A9WEI0 A0A1B0FQH9 T1IZW2 A0A2R5LBJ3 A0A131YZD2 A0A224Z0Y8 A0A293LZA7 A0A131XXK6 A0A1B6LU40 A0A2M4ALL9 L7MJP7 A0A2I9LNW1 A0A1E1X351 A0A3S3SBE5 A0A226F0T4 T1L1M9 A0A1B6IN68 X1WIF3 A0A0K8TMX6 A0A2S2Q883 A0A3B4BV38 A0A3P8VRE4 A0A3B3BGZ8 A0A0A1WLP4 A0A3B3T258 A0A2I4CWY0 M3XH89 A0A060X7M1
A0A2C9GPI7 A0A182VBG3 A0A2M3Z7E0 Q5TWP3 A0A023EVC7 A0A2M4AJZ7 A0A2M4AJR3 A0A1L8DMF9 A0A2M4CUU7 A0A2M4CUE5 A0A182YEH8 A0A1L8DMA7 A0A182QKK5 A0A182JB80 A0A2P8YYJ8 D6WR07 A0A151HYC6 E2BKT8 E2A0T0 A0A195ES80 A0A154PND9 A0A0L7RBJ3 A0A026WKM2 A0A151X559 A0A151IC13 F4X4T6 E9J4P7 A0A1Y1MER9 A0A088AJ90 A0A2A3ETZ5 A0A310S662 A0A1B6ECM8 A0A0A9YTN1 A0A0K8SCK0 A0A1B6EKV6 A0A1B6E8C9 J3JY04 A0A1W4WM14 Q9Y128 B4PG56 B3NFV1 A0A0K8U4A9 B4HJF4 A0A0J9RR46 A0A0A1WWK4 A0A034WQ32 B4L944 A0A1W4USF6 A0A0L0CAD6 B4LCA3 B4J205 B3M9P2 A0A1J1HZD1 A0A3B0KLS5 W8BMR1 Q29F82 A0A336MKJ2 A0A1A9UCS9 A0A1A9Z820 A0A0M4EKZ1 B4MKS1 E9FT92 A0A1A9Y4S9 A0A1B0BE36 E0VRU4 A0A1A9WEI0 A0A1B0FQH9 T1IZW2 A0A2R5LBJ3 A0A131YZD2 A0A224Z0Y8 A0A293LZA7 A0A131XXK6 A0A1B6LU40 A0A2M4ALL9 L7MJP7 A0A2I9LNW1 A0A1E1X351 A0A3S3SBE5 A0A226F0T4 T1L1M9 A0A1B6IN68 X1WIF3 A0A0K8TMX6 A0A2S2Q883 A0A3B4BV38 A0A3P8VRE4 A0A3B3BGZ8 A0A0A1WLP4 A0A3B3T258 A0A2I4CWY0 M3XH89 A0A060X7M1
Pubmed
26354079
20966253
12364791
14747013
17210077
24945155
+ More
25244985 29403074 18362917 19820115 20798317 24508170 30249741 21719571 21282665 28004739 25401762 26823975 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 22936249 25830018 25348373 26108605 24495485 15632085 18057021 21292972 20566863 26830274 28797301 25576852 29248469 28503490 26369729 24487278 29451363 29240929 9215903 24755649
25244985 29403074 18362917 19820115 20798317 24508170 30249741 21719571 21282665 28004739 25401762 26823975 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 22936249 25830018 25348373 26108605 24495485 15632085 18057021 21292972 20566863 26830274 28797301 25576852 29248469 28503490 26369729 24487278 29451363 29240929 9215903 24755649
EMBL
KQ459185
KPJ03286.1
KQ460772
KPJ12480.1
NWSH01001346
PCG71597.1
+ More
GGFJ01003683 MBW52824.1 APCN01000155 GGFM01003695 MBW24446.1 AAAB01008807 EAL41877.3 GAPW01000662 JAC12936.1 GGFK01007780 MBW41101.1 GGFK01007683 MBW41004.1 GFDF01006431 JAV07653.1 GGFL01004763 MBW68941.1 GGFL01004762 MBW68940.1 GFDF01006481 JAV07603.1 AXCN02000424 PYGN01000284 PSN49326.1 KQ971351 EFA06537.1 KQ976730 KYM76444.1 GL448826 EFN83687.1 GL435626 EFN72983.1 KQ981993 KYN31083.1 KQ434977 KZC12828.1 KQ414617 KOC68234.1 KK107198 QOIP01000007 EZA55629.1 RLU20314.1 KQ982522 KYQ55464.1 KQ978069 KYM97346.1 GL888679 EGI58509.1 GL768112 EFZ12193.1 GEZM01036286 JAV82825.1 KZ288185 PBC34974.1 KQ768636 OAD53087.1 GEDC01001600 JAS35698.1 GBHO01009161 JAG34443.1 GBRD01014809 GDHC01017079 JAG51017.1 JAQ01550.1 GECZ01031238 JAS38531.1 GEDC01003096 JAS34202.1 BT128127 AEE63088.1 AF145644 AE014296 AAD38619.1 AAF50454.1 AHN58003.1 CM000159 EDW93200.1 CH954178 EDV50713.1 GDHF01031174 JAI21140.1 CH480815 EDW40678.1 CM002912 KMY98336.1 GBXI01010853 JAD03439.1 GAKP01003084 JAC55868.1 CH933816 EDW17219.1 JRES01000681 KNC29221.1 CH940647 EDW68748.1 CH916366 EDV95930.1 CH902618 EDV39048.1 CVRI01000035 CRK92890.1 OUUW01000012 SPP87529.1 GAMC01006484 GAMC01006483 JAC00073.1 CH379067 EAL31249.2 UFQT01001407 SSX30415.1 CP012525 ALC44938.1 CH963847 EDW72777.2 KRF97632.1 GL732524 EFX89333.1 JXJN01012777 DS235562 EEB16100.1 CCAG010008844 JH431728 GGLE01002601 MBY06727.1 GEDV01005296 JAP83261.1 GFPF01011679 MAA22825.1 GFWV01009314 MAA34043.1 GEFM01004379 JAP71417.1 GEBQ01012793 JAT27184.1 GGFK01008350 MBW41671.1 GACK01000844 JAA64190.1 GFWZ01000082 MBW20072.1 GFAC01005478 JAT93710.1 NCKU01001105 RWS13073.1 LNIX01000001 OXA62811.1 CAEY01000920 GECU01019341 JAS88365.1 ABLF02037610 GDAI01002112 JAI15491.1 GGMS01004736 MBY73939.1 GBXI01014847 JAC99444.1 AFYH01173700 AFYH01173701 AFYH01173702 AFYH01173703 AFYH01173704 AFYH01173705 AFYH01173706 AFYH01173707 AFYH01173708 AFYH01173709 FR904892 CDQ73289.1
GGFJ01003683 MBW52824.1 APCN01000155 GGFM01003695 MBW24446.1 AAAB01008807 EAL41877.3 GAPW01000662 JAC12936.1 GGFK01007780 MBW41101.1 GGFK01007683 MBW41004.1 GFDF01006431 JAV07653.1 GGFL01004763 MBW68941.1 GGFL01004762 MBW68940.1 GFDF01006481 JAV07603.1 AXCN02000424 PYGN01000284 PSN49326.1 KQ971351 EFA06537.1 KQ976730 KYM76444.1 GL448826 EFN83687.1 GL435626 EFN72983.1 KQ981993 KYN31083.1 KQ434977 KZC12828.1 KQ414617 KOC68234.1 KK107198 QOIP01000007 EZA55629.1 RLU20314.1 KQ982522 KYQ55464.1 KQ978069 KYM97346.1 GL888679 EGI58509.1 GL768112 EFZ12193.1 GEZM01036286 JAV82825.1 KZ288185 PBC34974.1 KQ768636 OAD53087.1 GEDC01001600 JAS35698.1 GBHO01009161 JAG34443.1 GBRD01014809 GDHC01017079 JAG51017.1 JAQ01550.1 GECZ01031238 JAS38531.1 GEDC01003096 JAS34202.1 BT128127 AEE63088.1 AF145644 AE014296 AAD38619.1 AAF50454.1 AHN58003.1 CM000159 EDW93200.1 CH954178 EDV50713.1 GDHF01031174 JAI21140.1 CH480815 EDW40678.1 CM002912 KMY98336.1 GBXI01010853 JAD03439.1 GAKP01003084 JAC55868.1 CH933816 EDW17219.1 JRES01000681 KNC29221.1 CH940647 EDW68748.1 CH916366 EDV95930.1 CH902618 EDV39048.1 CVRI01000035 CRK92890.1 OUUW01000012 SPP87529.1 GAMC01006484 GAMC01006483 JAC00073.1 CH379067 EAL31249.2 UFQT01001407 SSX30415.1 CP012525 ALC44938.1 CH963847 EDW72777.2 KRF97632.1 GL732524 EFX89333.1 JXJN01012777 DS235562 EEB16100.1 CCAG010008844 JH431728 GGLE01002601 MBY06727.1 GEDV01005296 JAP83261.1 GFPF01011679 MAA22825.1 GFWV01009314 MAA34043.1 GEFM01004379 JAP71417.1 GEBQ01012793 JAT27184.1 GGFK01008350 MBW41671.1 GACK01000844 JAA64190.1 GFWZ01000082 MBW20072.1 GFAC01005478 JAT93710.1 NCKU01001105 RWS13073.1 LNIX01000001 OXA62811.1 CAEY01000920 GECU01019341 JAS88365.1 ABLF02037610 GDAI01002112 JAI15491.1 GGMS01004736 MBY73939.1 GBXI01014847 JAC99444.1 AFYH01173700 AFYH01173701 AFYH01173702 AFYH01173703 AFYH01173704 AFYH01173705 AFYH01173706 AFYH01173707 AFYH01173708 AFYH01173709 FR904892 CDQ73289.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000075902
UP000075882
UP000075840
+ More
UP000075903 UP000007062 UP000076408 UP000075886 UP000075880 UP000245037 UP000007266 UP000078540 UP000008237 UP000000311 UP000078541 UP000076502 UP000053825 UP000053097 UP000279307 UP000075809 UP000078542 UP000007755 UP000005203 UP000242457 UP000192223 UP000000803 UP000002282 UP000008711 UP000001292 UP000009192 UP000192221 UP000037069 UP000008792 UP000001070 UP000007801 UP000183832 UP000268350 UP000001819 UP000078200 UP000092445 UP000092553 UP000007798 UP000000305 UP000092443 UP000092460 UP000009046 UP000091820 UP000092444 UP000285301 UP000198287 UP000015104 UP000007819 UP000261440 UP000265120 UP000261560 UP000261540 UP000192220 UP000008672 UP000193380
UP000075903 UP000007062 UP000076408 UP000075886 UP000075880 UP000245037 UP000007266 UP000078540 UP000008237 UP000000311 UP000078541 UP000076502 UP000053825 UP000053097 UP000279307 UP000075809 UP000078542 UP000007755 UP000005203 UP000242457 UP000192223 UP000000803 UP000002282 UP000008711 UP000001292 UP000009192 UP000192221 UP000037069 UP000008792 UP000001070 UP000007801 UP000183832 UP000268350 UP000001819 UP000078200 UP000092445 UP000092553 UP000007798 UP000000305 UP000092443 UP000092460 UP000009046 UP000091820 UP000092444 UP000285301 UP000198287 UP000015104 UP000007819 UP000261440 UP000265120 UP000261560 UP000261540 UP000192220 UP000008672 UP000193380
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
ProteinModelPortal
A0A194QIK7
A0A0N1IDF5
A0A2A4JHR7
A0A2M4BII1
A0A182U1V8
A0A182KQA6
+ More
A0A2C9GPI7 A0A182VBG3 A0A2M3Z7E0 Q5TWP3 A0A023EVC7 A0A2M4AJZ7 A0A2M4AJR3 A0A1L8DMF9 A0A2M4CUU7 A0A2M4CUE5 A0A182YEH8 A0A1L8DMA7 A0A182QKK5 A0A182JB80 A0A2P8YYJ8 D6WR07 A0A151HYC6 E2BKT8 E2A0T0 A0A195ES80 A0A154PND9 A0A0L7RBJ3 A0A026WKM2 A0A151X559 A0A151IC13 F4X4T6 E9J4P7 A0A1Y1MER9 A0A088AJ90 A0A2A3ETZ5 A0A310S662 A0A1B6ECM8 A0A0A9YTN1 A0A0K8SCK0 A0A1B6EKV6 A0A1B6E8C9 J3JY04 A0A1W4WM14 Q9Y128 B4PG56 B3NFV1 A0A0K8U4A9 B4HJF4 A0A0J9RR46 A0A0A1WWK4 A0A034WQ32 B4L944 A0A1W4USF6 A0A0L0CAD6 B4LCA3 B4J205 B3M9P2 A0A1J1HZD1 A0A3B0KLS5 W8BMR1 Q29F82 A0A336MKJ2 A0A1A9UCS9 A0A1A9Z820 A0A0M4EKZ1 B4MKS1 E9FT92 A0A1A9Y4S9 A0A1B0BE36 E0VRU4 A0A1A9WEI0 A0A1B0FQH9 T1IZW2 A0A2R5LBJ3 A0A131YZD2 A0A224Z0Y8 A0A293LZA7 A0A131XXK6 A0A1B6LU40 A0A2M4ALL9 L7MJP7 A0A2I9LNW1 A0A1E1X351 A0A3S3SBE5 A0A226F0T4 T1L1M9 A0A1B6IN68 X1WIF3 A0A0K8TMX6 A0A2S2Q883 A0A3B4BV38 A0A3P8VRE4 A0A3B3BGZ8 A0A0A1WLP4 A0A3B3T258 A0A2I4CWY0 M3XH89 A0A060X7M1
A0A2C9GPI7 A0A182VBG3 A0A2M3Z7E0 Q5TWP3 A0A023EVC7 A0A2M4AJZ7 A0A2M4AJR3 A0A1L8DMF9 A0A2M4CUU7 A0A2M4CUE5 A0A182YEH8 A0A1L8DMA7 A0A182QKK5 A0A182JB80 A0A2P8YYJ8 D6WR07 A0A151HYC6 E2BKT8 E2A0T0 A0A195ES80 A0A154PND9 A0A0L7RBJ3 A0A026WKM2 A0A151X559 A0A151IC13 F4X4T6 E9J4P7 A0A1Y1MER9 A0A088AJ90 A0A2A3ETZ5 A0A310S662 A0A1B6ECM8 A0A0A9YTN1 A0A0K8SCK0 A0A1B6EKV6 A0A1B6E8C9 J3JY04 A0A1W4WM14 Q9Y128 B4PG56 B3NFV1 A0A0K8U4A9 B4HJF4 A0A0J9RR46 A0A0A1WWK4 A0A034WQ32 B4L944 A0A1W4USF6 A0A0L0CAD6 B4LCA3 B4J205 B3M9P2 A0A1J1HZD1 A0A3B0KLS5 W8BMR1 Q29F82 A0A336MKJ2 A0A1A9UCS9 A0A1A9Z820 A0A0M4EKZ1 B4MKS1 E9FT92 A0A1A9Y4S9 A0A1B0BE36 E0VRU4 A0A1A9WEI0 A0A1B0FQH9 T1IZW2 A0A2R5LBJ3 A0A131YZD2 A0A224Z0Y8 A0A293LZA7 A0A131XXK6 A0A1B6LU40 A0A2M4ALL9 L7MJP7 A0A2I9LNW1 A0A1E1X351 A0A3S3SBE5 A0A226F0T4 T1L1M9 A0A1B6IN68 X1WIF3 A0A0K8TMX6 A0A2S2Q883 A0A3B4BV38 A0A3P8VRE4 A0A3B3BGZ8 A0A0A1WLP4 A0A3B3T258 A0A2I4CWY0 M3XH89 A0A060X7M1
PDB
5JJD
E-value=9.95593e-57,
Score=559
Ontologies
GO
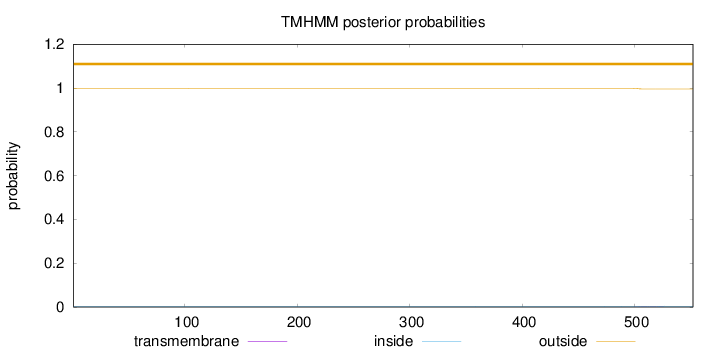
Topology
Length:
552
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05213
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00171
outside
1 - 552
Population Genetic Test Statistics
Pi
228.951432
Theta
178.777933
Tajima's D
1.230399
CLR
0.420707
CSRT
0.713964301784911
Interpretation
Uncertain
