Gene
KWMTBOMO11604
Pre Gene Modal
BGIBMGA004107
Annotation
PREDICTED:_high_affinity_cAMP-specific_and_IBMX-insensitive_3'?5'-cyclic_phosphodiesterase_8A_[Bombyx_mori]
Full name
Phosphodiesterase
+ More
High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8
High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8
Alternative Name
Phosphodiesterase 8
Location in the cell
Cytoplasmic Reliability : 1.136 Mitochondrial Reliability : 1.595
Sequence
CDS
ATGACACTAATGCAAACTGAAACAAAGGATATATGTTTTATGAAAACAGCAAGCCGCCGTCACGTTATAGACGACACGCTCTCTGACGCTTCGACTGCATATCTCAAAGATGAATTCGGTGTGCTGGCATATTTAGAGGCAGGATTCAACAGGGTAATGGTGGAAACAGTAAACACGACAGCTTGGTGCGCTGAAGTACTACAGGCGCGTTCCTCAGCCGCGTGCTCCCGGGCACAGATCGCAGCCACAGCCGCCTTGGCCGCTGCCGCCGACCGTTGCAGAGATTTGGTTGCTATCACTGACGACCAGCAGAGGATCCTCCTCACTAATAAATCCTGGTGCCGTATGCTGGGGTGGCGTTTGGAGGAAGGTCCGAGGCCAATCCACGAAGCGGCTGGAGGAGAAGCTCTACGACTGGCAACAACCGGAGCCAGGGACTGGGAGGCGCCAGTTACTCTCCGTCGCAGGGCTGCTCCAGATCCAATCATGCTGCCATGCAGGGGTGCCACTATAGGGCTCACTAAGACAACTTCTCACATAGTATTCGTCTGTGAGCCGAGCGCAGGAGAGGGCGACCGTGGTAGGGGCTCGCTGCACTCGATCAGACGCGGCTCGCTTGACGTGCGCTCAGTGGCCTCGGACGGACTGCGTCGAACCTCACTGGCCAAACTACAGGGACTACCTCTCGAAGCACCCATCACTAAGGTGATATCTCTACTATCGGCGGCTACGGAGGGTGCTCCGCCTTCCACCGTGGCCTGTATAGAGCGAGCCGTCGACATGCTCAGGACTTCAGAGCTGTACTCACCACAGATGAGAGACGAAACCAAACTCAGAGTCGAGGACCCCATAGCCACCGATCTCATTGGCGCACTGCTATCGGGAAATGCTAACGTCTTATCGTCGCGTCGAAGCAGCAATGACTCCACCGTCGTGCGTTCACAAACTAACCGAACCAACATCAAACACAAGACGACTGGTCAACTTCGGGAGCTTCTGGACACGGCTCTGAGCTGGGACTTCGAGATCTTCCGCCTGGAGGATCTGACCCGCGGACGGCCCCTCGCGCACCTCGGGCTCGCGCTCATGGGGGGCAGGTTCGACGTGTGCGCCGCCCTGGAGTGCGACGAGAGAACGCTCCTGCACTGGCTCACCGTCATCGAGACCAACTACCACGCCGTCAACACCTACCACAATTCCACGCATGCGGCAGATGTACTACAGGCAGTGGCGTACTTCCTGGAGAAGGATCGGCTGAAGAACCTTCTGGAGCCAGTGGAGGCGGCCGCGGCTCTGATCTCGGCGGCGGCGCACGACATCGACCACCCGGGCACCTCGTCCGCCTTCCTCTGCAACGCGCGACACCCGCTCGCGATCCTCTACAATGACGTCAGCGTGCTCGAGTCGCATCACGCCGCGCTCACATTCAAACTCACGCTCAACGACGACCGCGTGAACATATTCAAGAATCTCGACCGCGACACGTACAAGACGGTGCGGCACCACGTCATCGACATGATCCTCGCGACGGAGATGACGAAACACTTCGAGCATCTCGCCAAGTTCGTCAACGTGTTCTACGCTAAGTCCTCCGGTAGCAAGGAGGACGGCATGCACACTGATGAGCCATTATCACTAGACACCACAGCACTTTCTCAACCAGAGAATGTCTTGTTGGTGAAAAGAATGATGATCAAATGCGCTGATGTCTCGAACGCGTCACGACCTCAAAAGTTCGCTATGGAGTGGGCTAGACGGATTGCTGAGGAATATTTCCTACAGACGGATGAAGAAAAGGCGAAAGACTTGCCTGTCGTGATGCCAATGTTCGATCGAGCCACCTGCTCTATCCCGCGCTCACAAATCGGCTTCATCGACTACATCATCATAGACATGATGGAGGCCTGGGCTGCCTTCATCGACATGCCAGAACTAGTGAACCACGCCAGAAGCAACAACCAACACTGGCGGGAACTGGACGAGGCGGGAGTGACCACGCTCGCCGACGTCAAACGTGTTCAACGACAAAACGCTATACAAGCCCCTCCCACCACTACCGCCGAACCGACAACTGAAGAATAA
Protein
MTLMQTETKDICFMKTASRRHVIDDTLSDASTAYLKDEFGVLAYLEAGFNRVMVETVNTTAWCAEVLQARSSAACSRAQIAATAALAAAADRCRDLVAITDDQQRILLTNKSWCRMLGWRLEEGPRPIHEAAGGEALRLATTGARDWEAPVTLRRRAAPDPIMLPCRGATIGLTKTTSHIVFVCEPSAGEGDRGRGSLHSIRRGSLDVRSVASDGLRRTSLAKLQGLPLEAPITKVISLLSAATEGAPPSTVACIERAVDMLRTSELYSPQMRDETKLRVEDPIATDLIGALLSGNANVLSSRRSSNDSTVVRSQTNRTNIKHKTTGQLRELLDTALSWDFEIFRLEDLTRGRPLAHLGLALMGGRFDVCAALECDERTLLHWLTVIETNYHAVNTYHNSTHAADVLQAVAYFLEKDRLKNLLEPVEAAAALISAAAHDIDHPGTSSAFLCNARHPLAILYNDVSVLESHHAALTFKLTLNDDRVNIFKNLDRDTYKTVRHHVIDMILATEMTKHFEHLAKFVNVFYAKSSGSKEDGMHTDEPLSLDTTALSQPENVLLVKRMMIKCADVSNASRPQKFAMEWARRIAEEYFLQTDEEKAKDLPVVMPMFDRATCSIPRSQIGFIDYIIIDMMEAWAAFIDMPELVNHARSNNQHWRELDEAGVTTLADVKRVQRQNAIQAPPTTTAEPTTEE
Summary
Description
Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes (By similarity). Involved in the positive regulation of MAP kinase signaling and in inhibiting oxidative stress-induced cell death (PubMed:23509299).
Catalytic Activity
3',5'-cyclic AMP + H2O = AMP + H(+)
Cofactor
a divalent metal cation
Similarity
Belongs to the cyclic nucleotide phosphodiesterase family.
Belongs to the cyclic nucleotide phosphodiesterase family. PDE8 subfamily.
Belongs to the cyclic nucleotide phosphodiesterase family. PDE8 subfamily.
Keywords
Alternative splicing
cAMP
Complete proteome
Hydrolase
Metal-binding
Reference proteome
Feature
chain High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8
splice variant In isoform C.
splice variant In isoform C.
Uniprot
A0A194QCM1
A0A3S2NP76
A0A194QWB3
A0A212EGX9
A0A1Y1LB63
A0A1Y1LB55
+ More
A0A1Y1LGH2 A0A1B6C1Q8 A0A139WFF3 A0A139WF42 A0A1B6D166 N6TN97 A0A2H8TXF4 A0A2S2NVJ8 J9JTW7 A0A2S2QE64 A0A1B6GW39 A0A336LT50 A0A336LSU9 E0VRW3 E2C088 A0A1J1HIG6 A0A182WX29 A0A182LS27 A7URN7 A0A182UG75 A0A2J7QT83 A0A067RHJ8 A0A0K8TN27 Q7QJ01 A0A182PDQ2 Q16JF3 A0A182WJV1 A0A182RCM5 A0A158P0G0 A0A182GBB1 A0A182NJW4 A0A182R086 A0A151I4C0 A0A1S4FYS0 A0A0K8U7D3 A0A034VBY3 A0A0A1XJC2 E9IKR3 A0A0A1WUU5 W8BBA4 A0A1I8PV34 A0A0L0C916 A0A0A1XQ71 W8ARC4 A0A084VQY3 A0A182I2M1 B4NMM0 A0A182Y2P7 A0A195D1Y8 T1PKA6 A0A1I8MIV0 A0A1Q3FX90 A0A195E195 A0A195F2E0 A0A2M4A8P1 A0A182IU06 A0A2M4A7Z6 A0A151WXN6 Q28ZF2 A0A2M4CS56 A0A3B0JHX1 A0A1B6LW76 A0A1Q3FUP7 W5J8B3 A0A232FCU7 B3MFI3 T1PMT6 B4LLX8 A0A182FGD7 Q8MLR1 Q6NNF2-2 B3NPQ1 A0A026W1V4 A0A3L8D7R8 E1JGU6 K7IY17 B4P9Q8 F4WRM3 B4I8R1 Q6NNF2 A0A0J9U9F1 Q6NNF2-5 B4J7C7 A0A1W4UA72 A0A1W4UA69 A0A182KQI6 A0A182VP90 A0A1W4U9K6 A0A1W4U9K2 B4KS04 A0A0P6HD91
A0A1Y1LGH2 A0A1B6C1Q8 A0A139WFF3 A0A139WF42 A0A1B6D166 N6TN97 A0A2H8TXF4 A0A2S2NVJ8 J9JTW7 A0A2S2QE64 A0A1B6GW39 A0A336LT50 A0A336LSU9 E0VRW3 E2C088 A0A1J1HIG6 A0A182WX29 A0A182LS27 A7URN7 A0A182UG75 A0A2J7QT83 A0A067RHJ8 A0A0K8TN27 Q7QJ01 A0A182PDQ2 Q16JF3 A0A182WJV1 A0A182RCM5 A0A158P0G0 A0A182GBB1 A0A182NJW4 A0A182R086 A0A151I4C0 A0A1S4FYS0 A0A0K8U7D3 A0A034VBY3 A0A0A1XJC2 E9IKR3 A0A0A1WUU5 W8BBA4 A0A1I8PV34 A0A0L0C916 A0A0A1XQ71 W8ARC4 A0A084VQY3 A0A182I2M1 B4NMM0 A0A182Y2P7 A0A195D1Y8 T1PKA6 A0A1I8MIV0 A0A1Q3FX90 A0A195E195 A0A195F2E0 A0A2M4A8P1 A0A182IU06 A0A2M4A7Z6 A0A151WXN6 Q28ZF2 A0A2M4CS56 A0A3B0JHX1 A0A1B6LW76 A0A1Q3FUP7 W5J8B3 A0A232FCU7 B3MFI3 T1PMT6 B4LLX8 A0A182FGD7 Q8MLR1 Q6NNF2-2 B3NPQ1 A0A026W1V4 A0A3L8D7R8 E1JGU6 K7IY17 B4P9Q8 F4WRM3 B4I8R1 Q6NNF2 A0A0J9U9F1 Q6NNF2-5 B4J7C7 A0A1W4UA72 A0A1W4UA69 A0A182KQI6 A0A182VP90 A0A1W4U9K6 A0A1W4U9K2 B4KS04 A0A0P6HD91
EC Number
3.1.4.-
3.1.4.53
3.1.4.53
Pubmed
26354079
22118469
28004739
18362917
19820115
23537049
+ More
20566863 20798317 12364791 14747013 17210077 24845553 26369729 17510324 21347285 26483478 25348373 25830018 21282665 24495485 26108605 24438588 17994087 25244985 25315136 15632085 20920257 23761445 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15673286 23509299 24508170 30249741 20075255 17550304 21719571 22936249 20966253
20566863 20798317 12364791 14747013 17210077 24845553 26369729 17510324 21347285 26483478 25348373 25830018 21282665 24495485 26108605 24438588 17994087 25244985 25315136 15632085 20920257 23761445 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15673286 23509299 24508170 30249741 20075255 17550304 21719571 22936249 20966253
EMBL
KQ459185
KPJ03288.1
RSAL01000182
RVE44887.1
KQ461073
KPJ09594.1
+ More
AGBW02014996 OWR40747.1 GEZM01062028 JAV70111.1 GEZM01062023 JAV70118.1 GEZM01062027 GEZM01062024 JAV70117.1 GEDC01029994 JAS07304.1 KQ971354 KYB26565.1 KYB26564.1 GEDC01017839 JAS19459.1 APGK01058439 APGK01058440 APGK01058441 APGK01058442 APGK01058443 KB741288 ENN70710.1 GFXV01007170 MBW18975.1 GGMR01008463 MBY21082.1 ABLF02028341 GGMS01006816 MBY76019.1 GECZ01003152 JAS66617.1 UFQT01000164 SSX21065.1 SSX21064.1 DS235562 EEB16119.1 GL451753 EFN78640.1 CVRI01000001 CRK86262.1 AXCM01000307 AAAB01008807 EDO64678.1 NEVH01011198 PNF31799.1 KK852504 KDR22513.1 GDAI01002067 JAI15536.1 EAA04173.4 CH478011 EAT34419.1 ADTU01004819 ADTU01004820 ADTU01004821 ADTU01004822 JXUM01008983 JXUM01008984 JXUM01008985 JXUM01008986 JXUM01008987 JXUM01008988 KQ560273 KXJ83391.1 AXCN02000526 KQ976463 KYM84601.1 GDHF01029828 JAI22486.1 GAKP01019336 GAKP01019335 JAC39617.1 GBXI01003302 JAD10990.1 GL764026 EFZ18745.1 GBXI01011650 JAD02642.1 GAMC01019431 JAB87124.1 JRES01000729 KNC28923.1 GBXI01001272 JAD13020.1 GAMC01019432 JAB87123.1 ATLV01015348 ATLV01015349 ATLV01015350 KE525007 KFB40377.1 APCN01000169 CH964282 EDW85609.2 KQ976948 KYN06887.1 KA648495 AFP63124.1 GFDL01002870 JAV32175.1 KQ979824 KYN18908.1 KQ981856 KYN34745.1 GGFK01003853 MBW37174.1 GGFK01003602 MBW36923.1 KQ982656 KYQ52699.1 CM000071 EAL25661.4 GGFL01003965 MBW68143.1 OUUW01000001 SPP72989.1 GEBQ01012040 JAT27937.1 GFDL01003738 JAV31307.1 ADMH02002070 ETN59633.1 NNAY01000405 OXU28624.1 CH902619 EDV36668.2 KA650004 AFP64633.1 CH940648 EDW62001.2 AE013599 AAM68264.1 AY060327 BT021326 BT011336 BT133111 AAL25366.1 AAR96128.1 AAX33474.1 AEX98031.1 CH954179 EDV56842.2 KK107488 EZA50013.1 QOIP01000012 RLU15938.1 ACZ94550.2 CM000158 EDW92366.2 GL888291 EGI63168.1 CH480824 EDW56986.1 CM002911 KMY96080.1 CH916367 EDW01051.1 CH933808 EDW09445.2 GDIQ01021167 JAN73570.1
AGBW02014996 OWR40747.1 GEZM01062028 JAV70111.1 GEZM01062023 JAV70118.1 GEZM01062027 GEZM01062024 JAV70117.1 GEDC01029994 JAS07304.1 KQ971354 KYB26565.1 KYB26564.1 GEDC01017839 JAS19459.1 APGK01058439 APGK01058440 APGK01058441 APGK01058442 APGK01058443 KB741288 ENN70710.1 GFXV01007170 MBW18975.1 GGMR01008463 MBY21082.1 ABLF02028341 GGMS01006816 MBY76019.1 GECZ01003152 JAS66617.1 UFQT01000164 SSX21065.1 SSX21064.1 DS235562 EEB16119.1 GL451753 EFN78640.1 CVRI01000001 CRK86262.1 AXCM01000307 AAAB01008807 EDO64678.1 NEVH01011198 PNF31799.1 KK852504 KDR22513.1 GDAI01002067 JAI15536.1 EAA04173.4 CH478011 EAT34419.1 ADTU01004819 ADTU01004820 ADTU01004821 ADTU01004822 JXUM01008983 JXUM01008984 JXUM01008985 JXUM01008986 JXUM01008987 JXUM01008988 KQ560273 KXJ83391.1 AXCN02000526 KQ976463 KYM84601.1 GDHF01029828 JAI22486.1 GAKP01019336 GAKP01019335 JAC39617.1 GBXI01003302 JAD10990.1 GL764026 EFZ18745.1 GBXI01011650 JAD02642.1 GAMC01019431 JAB87124.1 JRES01000729 KNC28923.1 GBXI01001272 JAD13020.1 GAMC01019432 JAB87123.1 ATLV01015348 ATLV01015349 ATLV01015350 KE525007 KFB40377.1 APCN01000169 CH964282 EDW85609.2 KQ976948 KYN06887.1 KA648495 AFP63124.1 GFDL01002870 JAV32175.1 KQ979824 KYN18908.1 KQ981856 KYN34745.1 GGFK01003853 MBW37174.1 GGFK01003602 MBW36923.1 KQ982656 KYQ52699.1 CM000071 EAL25661.4 GGFL01003965 MBW68143.1 OUUW01000001 SPP72989.1 GEBQ01012040 JAT27937.1 GFDL01003738 JAV31307.1 ADMH02002070 ETN59633.1 NNAY01000405 OXU28624.1 CH902619 EDV36668.2 KA650004 AFP64633.1 CH940648 EDW62001.2 AE013599 AAM68264.1 AY060327 BT021326 BT011336 BT133111 AAL25366.1 AAR96128.1 AAX33474.1 AEX98031.1 CH954179 EDV56842.2 KK107488 EZA50013.1 QOIP01000012 RLU15938.1 ACZ94550.2 CM000158 EDW92366.2 GL888291 EGI63168.1 CH480824 EDW56986.1 CM002911 KMY96080.1 CH916367 EDW01051.1 CH933808 EDW09445.2 GDIQ01021167 JAN73570.1
Proteomes
UP000053268
UP000283053
UP000053240
UP000007151
UP000007266
UP000019118
+ More
UP000007819 UP000009046 UP000008237 UP000183832 UP000076407 UP000075883 UP000007062 UP000075902 UP000235965 UP000027135 UP000075885 UP000008820 UP000075920 UP000075900 UP000005205 UP000069940 UP000249989 UP000075884 UP000075886 UP000078540 UP000095300 UP000037069 UP000030765 UP000075840 UP000007798 UP000076408 UP000078542 UP000095301 UP000078492 UP000078541 UP000075880 UP000075809 UP000001819 UP000268350 UP000000673 UP000215335 UP000007801 UP000008792 UP000069272 UP000000803 UP000008711 UP000053097 UP000279307 UP000002358 UP000002282 UP000007755 UP000001292 UP000001070 UP000192221 UP000075882 UP000009192
UP000007819 UP000009046 UP000008237 UP000183832 UP000076407 UP000075883 UP000007062 UP000075902 UP000235965 UP000027135 UP000075885 UP000008820 UP000075920 UP000075900 UP000005205 UP000069940 UP000249989 UP000075884 UP000075886 UP000078540 UP000095300 UP000037069 UP000030765 UP000075840 UP000007798 UP000076408 UP000078542 UP000095301 UP000078492 UP000078541 UP000075880 UP000075809 UP000001819 UP000268350 UP000000673 UP000215335 UP000007801 UP000008792 UP000069272 UP000000803 UP000008711 UP000053097 UP000279307 UP000002358 UP000002282 UP000007755 UP000001292 UP000001070 UP000192221 UP000075882 UP000009192
Interpro
Gene 3D
ProteinModelPortal
A0A194QCM1
A0A3S2NP76
A0A194QWB3
A0A212EGX9
A0A1Y1LB63
A0A1Y1LB55
+ More
A0A1Y1LGH2 A0A1B6C1Q8 A0A139WFF3 A0A139WF42 A0A1B6D166 N6TN97 A0A2H8TXF4 A0A2S2NVJ8 J9JTW7 A0A2S2QE64 A0A1B6GW39 A0A336LT50 A0A336LSU9 E0VRW3 E2C088 A0A1J1HIG6 A0A182WX29 A0A182LS27 A7URN7 A0A182UG75 A0A2J7QT83 A0A067RHJ8 A0A0K8TN27 Q7QJ01 A0A182PDQ2 Q16JF3 A0A182WJV1 A0A182RCM5 A0A158P0G0 A0A182GBB1 A0A182NJW4 A0A182R086 A0A151I4C0 A0A1S4FYS0 A0A0K8U7D3 A0A034VBY3 A0A0A1XJC2 E9IKR3 A0A0A1WUU5 W8BBA4 A0A1I8PV34 A0A0L0C916 A0A0A1XQ71 W8ARC4 A0A084VQY3 A0A182I2M1 B4NMM0 A0A182Y2P7 A0A195D1Y8 T1PKA6 A0A1I8MIV0 A0A1Q3FX90 A0A195E195 A0A195F2E0 A0A2M4A8P1 A0A182IU06 A0A2M4A7Z6 A0A151WXN6 Q28ZF2 A0A2M4CS56 A0A3B0JHX1 A0A1B6LW76 A0A1Q3FUP7 W5J8B3 A0A232FCU7 B3MFI3 T1PMT6 B4LLX8 A0A182FGD7 Q8MLR1 Q6NNF2-2 B3NPQ1 A0A026W1V4 A0A3L8D7R8 E1JGU6 K7IY17 B4P9Q8 F4WRM3 B4I8R1 Q6NNF2 A0A0J9U9F1 Q6NNF2-5 B4J7C7 A0A1W4UA72 A0A1W4UA69 A0A182KQI6 A0A182VP90 A0A1W4U9K6 A0A1W4U9K2 B4KS04 A0A0P6HD91
A0A1Y1LGH2 A0A1B6C1Q8 A0A139WFF3 A0A139WF42 A0A1B6D166 N6TN97 A0A2H8TXF4 A0A2S2NVJ8 J9JTW7 A0A2S2QE64 A0A1B6GW39 A0A336LT50 A0A336LSU9 E0VRW3 E2C088 A0A1J1HIG6 A0A182WX29 A0A182LS27 A7URN7 A0A182UG75 A0A2J7QT83 A0A067RHJ8 A0A0K8TN27 Q7QJ01 A0A182PDQ2 Q16JF3 A0A182WJV1 A0A182RCM5 A0A158P0G0 A0A182GBB1 A0A182NJW4 A0A182R086 A0A151I4C0 A0A1S4FYS0 A0A0K8U7D3 A0A034VBY3 A0A0A1XJC2 E9IKR3 A0A0A1WUU5 W8BBA4 A0A1I8PV34 A0A0L0C916 A0A0A1XQ71 W8ARC4 A0A084VQY3 A0A182I2M1 B4NMM0 A0A182Y2P7 A0A195D1Y8 T1PKA6 A0A1I8MIV0 A0A1Q3FX90 A0A195E195 A0A195F2E0 A0A2M4A8P1 A0A182IU06 A0A2M4A7Z6 A0A151WXN6 Q28ZF2 A0A2M4CS56 A0A3B0JHX1 A0A1B6LW76 A0A1Q3FUP7 W5J8B3 A0A232FCU7 B3MFI3 T1PMT6 B4LLX8 A0A182FGD7 Q8MLR1 Q6NNF2-2 B3NPQ1 A0A026W1V4 A0A3L8D7R8 E1JGU6 K7IY17 B4P9Q8 F4WRM3 B4I8R1 Q6NNF2 A0A0J9U9F1 Q6NNF2-5 B4J7C7 A0A1W4UA72 A0A1W4UA69 A0A182KQI6 A0A182VP90 A0A1W4U9K6 A0A1W4U9K2 B4KS04 A0A0P6HD91
PDB
3ECN
E-value=1.62042e-109,
Score=1015
Ontologies
PATHWAY
GO
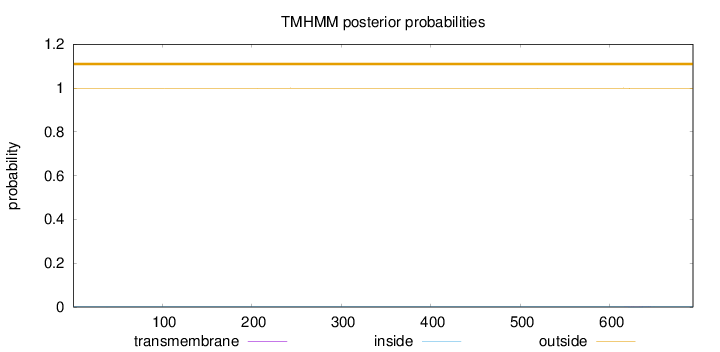
Topology
Length:
693
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0287
Exp number, first 60 AAs:
0.00112
Total prob of N-in:
0.00081
outside
1 - 693
Population Genetic Test Statistics
Pi
245.481875
Theta
167.966071
Tajima's D
1.381901
CLR
0.250323
CSRT
0.757562121893905
Interpretation
Uncertain
