Gene
KWMTBOMO11600
Pre Gene Modal
BGIBMGA004104
Annotation
PREDICTED:_protein_dead_ringer_isoform_X2_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.458
Sequence
CDS
ATGGCCAAGTCGGTCCTAGACCTGTACGAGCTGTACAACCTTGTGATAGCACGTGGAGGGCTCGTGGAGGTGATCAACAAGAAACTCTGGCAGGAGATAATTAAGGGACTCCGGCTGCCGTCTTCTATCACGTCCGCTGCCTTCACTTTACGCACTCAATATATGAAATATCTATACGACTATGAATGTGAGAAGAAAAATCTGTCCACGCGCAGCGAGTTGGATGCTGCTATTGAAGGTAACAAGCGCGAGGGAAGACGCGCCTCCGGACAATACGACGCCCAAGCTGCTTTAGCAATGCCGCCATTGAATCGCGGAGTCCCGGCCTCTCTCGCCCAGCTCTCCCAGCACATGCAGCCGCTCTCTCTCTCCCTCGGAGGAGGAGTCGCTGGAGTACCACGTCTACCACCACTGCCTCCACATCCCACTCACATCTCACAACACGATATTGAATACAGAGTAAGGGAATATATGAAGATGATACAACAACAGCGGGAGATGATGAGGAATGGTTCAGAATCGCCACCAGGCGGTAACGGGCCCACTCCTATGATCTCGCCGCGTGATGCCCTTAACGCTATTGATGTGTCTCGACTGACCCTTTGGTCTCTGTACAATAACAACAACCATAGCCCGCAGCCGGAACTAGAACCGCAACGAGAGGCCTTGAACCTTAGCGAGAGTCCGAATTCTATCACCAGCGGCTGCAAGCGTGAAGCGTGCCGGTCTCCCGCGCCGCCCCCCGCCAAGCGCCGCACCCCGCGCCCGCCCTCCCCGCGCCTCCCCTCCCCGCCGCGTCAGCCCTGCAGCAACACACCCCAGGTCAACGGCTCCAACGGAATGAACGGGGCTGCTGCCTTTAAAATCACTACTAAAGGTGACGCCAGCACTGGGGACCAGCAACTCGTAGTCTCCATCGAACTGAATGGCGTCACGTACGAAGGCGTATTGTTCCCATCGCAAAACAGCAACGGTCAGAACGGGCACAGGCAAATGGTGTCCTAG
Protein
MAKSVLDLYELYNLVIARGGLVEVINKKLWQEIIKGLRLPSSITSAAFTLRTQYMKYLYDYECEKKNLSTRSELDAAIEGNKREGRRASGQYDAQAALAMPPLNRGVPASLAQLSQHMQPLSLSLGGGVAGVPRLPPLPPHPTHISQHDIEYRVREYMKMIQQQREMMRNGSESPPGGNGPTPMISPRDALNAIDVSRLTLWSLYNNNNHSPQPELEPQREALNLSESPNSITSGCKREACRSPAPPPAKRRTPRPPSPRLPSPPRQPCSNTPQVNGSNGMNGAAAFKITTKGDASTGDQQLVVSIELNGVTYEGVLFPSQNSNGQNGHRQMVS
Summary
Uniprot
H9J3L7
A0A212EW44
A0A3S2TJE0
A0A2H1VJH4
A0A2A4JMV4
A0A194QIL2
+ More
A0A194QVN6 S4PDQ6 A0A139WF56 A0A139WFB1 A0A1Y1JZG9 A0A1Y1K447 A0A1Y1JWE4 A0A0C9RE32 U5EZ56 A0A0P6IJP4 A0A1I8P704 A0A023EY42 A0A1I8MN97 A0A1B6JUY7 A0A1J1J922 A0A182U1C7 A0A1W4WJE4 A0A182VEX8 A0A182LBG7 A7UT87 A0A2M4CLI2 A0A182ITE9 A0A084W2F0 A0A1Q3FW38 A0A1Q3FVK3 A0A2M4AAP6 A0A0M4EV64 A0A1B6DEC3 A0A0L0C9N2 B4NMM1 A0A0R3NTT2 B4P9Q7 A0A0R1DVG2 A0A0P6FRC2 A0A0J9RJU9 A0A0J9RJ83 B4I8R0 B4GHU8 Q28ZF1 A0A0R3NNF4 A0A0Q5VQ35 B3NPQ0 A0A0Q5VP34 A0A0Q5VW04 A0A1B0AMK3 A0A1A9XH89 B4LLY0 A0A1B0FDL6 A0A0Q9X8M2 A0A0Q9X911 B4KS05 A0A1B6M9L7
A0A194QVN6 S4PDQ6 A0A139WF56 A0A139WFB1 A0A1Y1JZG9 A0A1Y1K447 A0A1Y1JWE4 A0A0C9RE32 U5EZ56 A0A0P6IJP4 A0A1I8P704 A0A023EY42 A0A1I8MN97 A0A1B6JUY7 A0A1J1J922 A0A182U1C7 A0A1W4WJE4 A0A182VEX8 A0A182LBG7 A7UT87 A0A2M4CLI2 A0A182ITE9 A0A084W2F0 A0A1Q3FW38 A0A1Q3FVK3 A0A2M4AAP6 A0A0M4EV64 A0A1B6DEC3 A0A0L0C9N2 B4NMM1 A0A0R3NTT2 B4P9Q7 A0A0R1DVG2 A0A0P6FRC2 A0A0J9RJU9 A0A0J9RJ83 B4I8R0 B4GHU8 Q28ZF1 A0A0R3NNF4 A0A0Q5VQ35 B3NPQ0 A0A0Q5VP34 A0A0Q5VW04 A0A1B0AMK3 A0A1A9XH89 B4LLY0 A0A1B0FDL6 A0A0Q9X8M2 A0A0Q9X911 B4KS05 A0A1B6M9L7
Pubmed
EMBL
BABH01021584
AGBW02012080
OWR45718.1
RSAL01000096
RVE47701.1
ODYU01002724
+ More
SOQ40582.1 NWSH01001076 PCG72730.1 KQ459185 KPJ03291.1 KQ461073 KPJ09598.1 GAIX01004852 JAA87708.1 KQ971354 KYB26562.1 KYB26561.1 GEZM01098929 JAV53638.1 GEZM01098930 JAV53637.1 GEZM01098928 JAV53639.1 GBYB01014819 JAG84586.1 GANO01001398 JAB58473.1 GDIQ01003326 JAN91411.1 GAPW01000139 JAC13459.1 GECU01004707 JAT03000.1 CVRI01000075 CRL08554.1 AAAB01008900 EDO64092.1 GGFL01001965 MBW66143.1 ATLV01019610 ATLV01019611 ATLV01019612 ATLV01019613 KE525275 KFB44394.1 GFDL01003412 JAV31633.1 GFDL01003411 JAV31634.1 GGFK01004535 MBW37856.1 CP012524 ALC41712.1 GEDC01013308 JAS23990.1 JRES01000729 KNC28915.1 CH964282 EDW85610.1 CM000071 KRT02437.1 CM000158 EDW92365.2 KRK00478.1 GDIQ01045405 JAN49332.1 CM002911 KMY96077.1 KMY96078.1 CH480824 EDW56985.1 CH479183 EDW36068.1 EAL25662.2 KRT02436.1 CH954179 KQS63044.1 EDV56841.2 KQS63043.1 KQS63045.1 JXJN01000483 CH940648 EDW62003.2 CCAG010008323 CH933808 KRG04668.1 KRG04669.1 EDW09446.2 GEBQ01007345 JAT32632.1
SOQ40582.1 NWSH01001076 PCG72730.1 KQ459185 KPJ03291.1 KQ461073 KPJ09598.1 GAIX01004852 JAA87708.1 KQ971354 KYB26562.1 KYB26561.1 GEZM01098929 JAV53638.1 GEZM01098930 JAV53637.1 GEZM01098928 JAV53639.1 GBYB01014819 JAG84586.1 GANO01001398 JAB58473.1 GDIQ01003326 JAN91411.1 GAPW01000139 JAC13459.1 GECU01004707 JAT03000.1 CVRI01000075 CRL08554.1 AAAB01008900 EDO64092.1 GGFL01001965 MBW66143.1 ATLV01019610 ATLV01019611 ATLV01019612 ATLV01019613 KE525275 KFB44394.1 GFDL01003412 JAV31633.1 GFDL01003411 JAV31634.1 GGFK01004535 MBW37856.1 CP012524 ALC41712.1 GEDC01013308 JAS23990.1 JRES01000729 KNC28915.1 CH964282 EDW85610.1 CM000071 KRT02437.1 CM000158 EDW92365.2 KRK00478.1 GDIQ01045405 JAN49332.1 CM002911 KMY96077.1 KMY96078.1 CH480824 EDW56985.1 CH479183 EDW36068.1 EAL25662.2 KRT02436.1 CH954179 KQS63044.1 EDV56841.2 KQS63043.1 KQS63045.1 JXJN01000483 CH940648 EDW62003.2 CCAG010008323 CH933808 KRG04668.1 KRG04669.1 EDW09446.2 GEBQ01007345 JAT32632.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000053268
UP000053240
+ More
UP000007266 UP000095300 UP000095301 UP000183832 UP000075902 UP000192223 UP000075903 UP000075882 UP000007062 UP000075880 UP000030765 UP000092553 UP000037069 UP000007798 UP000001819 UP000002282 UP000001292 UP000008744 UP000008711 UP000092460 UP000092443 UP000008792 UP000092444 UP000009192
UP000007266 UP000095300 UP000095301 UP000183832 UP000075902 UP000192223 UP000075903 UP000075882 UP000007062 UP000075880 UP000030765 UP000092553 UP000037069 UP000007798 UP000001819 UP000002282 UP000001292 UP000008744 UP000008711 UP000092460 UP000092443 UP000008792 UP000092444 UP000009192
PRIDE
Pfam
PF01388 ARID
SUPFAM
SSF46774
SSF46774
Gene 3D
ProteinModelPortal
H9J3L7
A0A212EW44
A0A3S2TJE0
A0A2H1VJH4
A0A2A4JMV4
A0A194QIL2
+ More
A0A194QVN6 S4PDQ6 A0A139WF56 A0A139WFB1 A0A1Y1JZG9 A0A1Y1K447 A0A1Y1JWE4 A0A0C9RE32 U5EZ56 A0A0P6IJP4 A0A1I8P704 A0A023EY42 A0A1I8MN97 A0A1B6JUY7 A0A1J1J922 A0A182U1C7 A0A1W4WJE4 A0A182VEX8 A0A182LBG7 A7UT87 A0A2M4CLI2 A0A182ITE9 A0A084W2F0 A0A1Q3FW38 A0A1Q3FVK3 A0A2M4AAP6 A0A0M4EV64 A0A1B6DEC3 A0A0L0C9N2 B4NMM1 A0A0R3NTT2 B4P9Q7 A0A0R1DVG2 A0A0P6FRC2 A0A0J9RJU9 A0A0J9RJ83 B4I8R0 B4GHU8 Q28ZF1 A0A0R3NNF4 A0A0Q5VQ35 B3NPQ0 A0A0Q5VP34 A0A0Q5VW04 A0A1B0AMK3 A0A1A9XH89 B4LLY0 A0A1B0FDL6 A0A0Q9X8M2 A0A0Q9X911 B4KS05 A0A1B6M9L7
A0A194QVN6 S4PDQ6 A0A139WF56 A0A139WFB1 A0A1Y1JZG9 A0A1Y1K447 A0A1Y1JWE4 A0A0C9RE32 U5EZ56 A0A0P6IJP4 A0A1I8P704 A0A023EY42 A0A1I8MN97 A0A1B6JUY7 A0A1J1J922 A0A182U1C7 A0A1W4WJE4 A0A182VEX8 A0A182LBG7 A7UT87 A0A2M4CLI2 A0A182ITE9 A0A084W2F0 A0A1Q3FW38 A0A1Q3FVK3 A0A2M4AAP6 A0A0M4EV64 A0A1B6DEC3 A0A0L0C9N2 B4NMM1 A0A0R3NTT2 B4P9Q7 A0A0R1DVG2 A0A0P6FRC2 A0A0J9RJU9 A0A0J9RJ83 B4I8R0 B4GHU8 Q28ZF1 A0A0R3NNF4 A0A0Q5VQ35 B3NPQ0 A0A0Q5VP34 A0A0Q5VW04 A0A1B0AMK3 A0A1A9XH89 B4LLY0 A0A1B0FDL6 A0A0Q9X8M2 A0A0Q9X911 B4KS05 A0A1B6M9L7
PDB
1KQQ
E-value=1.02884e-38,
Score=401
Ontologies
GO
Topology
Subcellular location
Nucleus
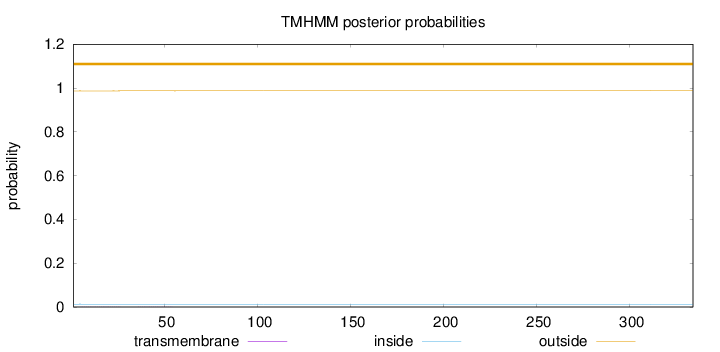
Length:
334
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01516
Exp number, first 60 AAs:
0.0146
Total prob of N-in:
0.01304
outside
1 - 334
Population Genetic Test Statistics
Pi
213.653026
Theta
172.175117
Tajima's D
0.795547
CLR
0.534386
CSRT
0.6021198940053
Interpretation
Uncertain
