Gene
KWMTBOMO11595
Pre Gene Modal
BGIBMGA004017
Annotation
PREDICTED:_GPI_ethanolamine_phosphate_transferase_2_isoform_X1_[Bombyx_mori]
Full name
GPI ethanolamine phosphate transferase 2
Alternative Name
GPI7 homolog
Phosphatidylinositol-glycan biosynthesis class G protein
Phosphatidylinositol-glycan biosynthesis class G protein
Location in the cell
PlasmaMembrane Reliability : 4.787
Sequence
CDS
ATGATCACATTATTGCAATACCGATGGACAGGGCTCTTCCTTCTAATCACGGCCTTAGGAGCATTAACCCTTTTCCTGTATGGATTTTTTCCACTAAAATATCATTCAGGAAAATTTGCCCACATGAGTGACCTTCCTAACTTCATAGATGGTGTAAGTATTGACGGACAGCAAGTATATAACTCTGGGGAGAATAGTGTGATTCTAATGGTTATTGATGGATTACGCTATGATTTTGTAACAGAAGAATATATGGCATATACAGGCCAACTCTTGAAGAACAAATCAGCTTGCATTTATGTTACTGTTGCTGAGCCTCCTACTGTGACTATGCCCAGAATAAAATCACCAGAACGGAAAAAGGAAAAAGCACAGTGGAGACGCACAATAAACTTCTGGAGGGACATATTCGCCATGATGACTGGGAGTGTATCGACGTTTGCCGATGTAGCGTTAAACTTCGGAGCGCCTGCTGTTCGAGGGGACAGCGTACTACGCGTCGCCGCTTCTAGAGGTCGACGCACCGTCATGTACGGGGACGACACCTGGATCAGACTATTCCCAGGCCTGTGGTCCGAATACGACGGAACCACTTCGTTCTTCGTGACCGATTATACAGAGGTGGACAACAATGTCACCAGGCATTTAGACAAAGTGTTGGCGCCTGATGAAAAGAAACAACCGAAATTCGACCTTTTAGTACTGCACTATTTAGGCTTAGATCACATTGGTCACTTGGAAGGCGCCCGCAGTACAAAAATCAAACCGAAGCTAGTGGAAATGGATGAGATAGTGAAGAAGATTTATGTGGCCATGCAAAAATGGGATCGTAACGGAATCCTAATAGTTTGCGGAGACCACGGCATGCGGGACGCGGGGGGCCACGGGGGCTCCACTCCCGCAGAGGTTCTAGTTCCTCTGGTGGTCATAAAAAGTCAAGATTTCGAATGCCCACATCCATTAGGGCCAGGACCTACGGTGGCTCAGGTCGACGTTGCGCCCACGATATCCTGGTTGCTAAACGCCCCGCCCCCCGGGGATAGCACGGGCCGGATACTGCCGTCCCTGCTCCCCGCCGACACGAGACAGCATCTCTACTTACTTCACATCATAGCTGAAAGAATGGCAAAACAAAACGACTTGCCCACAGATAAAGAGTTTTACAAGCAATTCGAGAGGGCCGAAAATCAATTTGCCCACTATTTGGCGACCGGTCAGCAAAGCGCCGCGAAAATATCAAAAGAGTTCTACGAACAGTCGCTGGCGGAGATGACCGAGTACCTCAACGGAACCACGGTCGACTTCGACATGTTCTCTATAGTAGTTGCTATAATACTGCTTTATTTTATTGTCGCGTCGCTTCTGTGCGTAACTCTATACTCACTCCAGCCGGACCACAGCAGGAAACACAGTACGACGAAGCGAGGCTACCAAAGCTCTAACGCCGGTAGAATAGTGGCGTTTCTATTCATGCTCTGCTTGTCCGGTAGCGGCTTGATCGTCGCCTGTTTCATAACCGAAACCAAAAGTCAGCTGTGCTCGTTCAACGGGCTCTGGCTACCGGTCCTTTTAGTCGTCGCGGCAGCGTTCACAATAACATACTTCATCATGAAAACCGGCATCGAGAGAATAAAAAATCTGTCGTCGCTAAACGAGCTGAACGCGATCGATTTTCTACTGATCGGCGGGACAGTTTTCCATGCGTGGTCTTTCTTCGGCACGAGTTTCATCGAGGAAGAGCACATGACGTGGTATTTCTTCTGGAACACATTCATGTTTTTCGTTTTGATACGGAGCATCGTGGTTGTCGTTATGTATTGGAGTAAGAAGATGACCGGCGCGACTGAAGTACAAGAGAAACCTGGCTTAGATGAGAAAATGAGCGCCGTCGGTATCAGTATTGTCCCGAAATGGGTCCTACTCATAGCCTTACATAGATATTTGCGCACCATGAACCAGACCGGGGACCGCTGGTTGTTCCTGCCGGACACAGCGGACTGGCTCAACGAACCTGAGAACTCGCTATACTTAGAACTCCACGTGATCATTGGAACTATAATGACAGTATTAATATGTGTCCACAATCTCGTTTACACGAATAACTTGATGTACATCCACACGGCGCTGACGGCCGCGGCCGCGACCTGCACCTTCTGCTACAGGGTTGCCACGCACGCCGTGCACGCACCCTTCGAGGACATGAAGACCTGGGACCCCATTATAATCGTCAGCGTTTTCTGGGGAATCCTAATGGCCCAGTTCATATACGAGATCATCACTTACGTTGGATTCTTCAAAAGCTGCGGCGTCCATCCCGCCGAGCACGCAATGAACAGCAAAAAGTTCGATTACGTGAAACCTAAGACAAAAACTGAAGATTATATCTACGATCCGACAATTGAGGAGCCGTGGAACGTCGAGGAAGTCAAACTGAATCTCGCTCGTTCGCTTTCGCATCTGATGCTCAATAATATGATGCTAGTAATACTGTTGCTGATGAGACCGCACAACGTGGTAATGGTGCCCAGTGTATACATTACGTGTGTCGTGAGTTCGAAATGTGTGGACCATAAACTGCTGGATCGGAGGTCCGCTAGGAACACGGACGTCGCGGACGTTCTGAGCAGGACGCTGGTGCACATGTGGATCGGGATACTGTTCTTCTTCTATCAGGGTAATTCGAACAGTCTGGCGTCTGTGGACCTGGGGTCGGGGTATGTTGGTCTGCGCGAGTACAGCCCTGTGAGGGTGGCGACGCGCATGGGGCTCCACGCGTACGCCGGACCCGCGCTGGCTGGCGCCGTGCTGTTCTGCACCCTCGCTGCGGAGGCCCGCGATTGGCAACAGTACTTAAAATCAATATGGCGGGCTACAAACATATTGGCGCTCCACAGAGTATACGCTGTCGTCTTATATACAGTGATAGCGACAATATTTCGACACCATTTATTTGTGTGGTCTGTCTTCTCTCCTAAACTATTATACGATTTCGTTGCAACAGTCTTTGGTGTACAGGCGTTAGCTACCATAGGTTATATTATTTTTTTAACGCATGTTACGAGTTGGCTGGCTAGAGTACTAATCACTAGATCAGGACTGAGGTAA
Protein
MITLLQYRWTGLFLLITALGALTLFLYGFFPLKYHSGKFAHMSDLPNFIDGVSIDGQQVYNSGENSVILMVIDGLRYDFVTEEYMAYTGQLLKNKSACIYVTVAEPPTVTMPRIKSPERKKEKAQWRRTINFWRDIFAMMTGSVSTFADVALNFGAPAVRGDSVLRVAASRGRRTVMYGDDTWIRLFPGLWSEYDGTTSFFVTDYTEVDNNVTRHLDKVLAPDEKKQPKFDLLVLHYLGLDHIGHLEGARSTKIKPKLVEMDEIVKKIYVAMQKWDRNGILIVCGDHGMRDAGGHGGSTPAEVLVPLVVIKSQDFECPHPLGPGPTVAQVDVAPTISWLLNAPPPGDSTGRILPSLLPADTRQHLYLLHIIAERMAKQNDLPTDKEFYKQFERAENQFAHYLATGQQSAAKISKEFYEQSLAEMTEYLNGTTVDFDMFSIVVAIILLYFIVASLLCVTLYSLQPDHSRKHSTTKRGYQSSNAGRIVAFLFMLCLSGSGLIVACFITETKSQLCSFNGLWLPVLLVVAAAFTITYFIMKTGIERIKNLSSLNELNAIDFLLIGGTVFHAWSFFGTSFIEEEHMTWYFFWNTFMFFVLIRSIVVVVMYWSKKMTGATEVQEKPGLDEKMSAVGISIVPKWVLLIALHRYLRTMNQTGDRWLFLPDTADWLNEPENSLYLELHVIIGTIMTVLICVHNLVYTNNLMYIHTALTAAAATCTFCYRVATHAVHAPFEDMKTWDPIIIVSVFWGILMAQFIYEIITYVGFFKSCGVHPAEHAMNSKKFDYVKPKTKTEDYIYDPTIEEPWNVEEVKLNLARSLSHLMLNNMMLVILLLMRPHNVVMVPSVYITCVVSSKCVDHKLLDRRSARNTDVADVLSRTLVHMWIGILFFFYQGNSNSLASVDLGSGYVGLREYSPVRVATRMGLHAYAGPALAGAVLFCTLAAEARDWQQYLKSIWRATNILALHRVYAVVLYTVIATIFRHHLFVWSVFSPKLLYDFVATVFGVQALATIGYIIFLTHVTSWLARVLITRSGLR
Summary
Description
Ethanolamine phosphate transferase involved in glycosylphosphatidylinositol-anchor biosynthesis. Transfers ethanolamine phosphate to the GPI second mannose.
Subunit
Forms a complex with PIGF. PIGF is required to stabilize it. Competes with PIGO for the binding of PIGF.
Similarity
Belongs to the PIGG/PIGN/PIGO family. PIGG subfamily.
Keywords
Alternative splicing
Complete proteome
Disease mutation
Endoplasmic reticulum
Glycoprotein
GPI-anchor biosynthesis
Membrane
Mental retardation
Polymorphism
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Feature
chain GPI ethanolamine phosphate transferase 2
splice variant In isoform 6.
sequence variant In dbSNP:rs34120878.
splice variant In isoform 6.
sequence variant In dbSNP:rs34120878.
Uniprot
A0A2H1V5Z2
A0A3S2LZI0
A0A2A4JCS8
A0A194QIL6
A0A194QWC4
H9J3D0
+ More
A0A2H1WSG9 A0A212EW94 A0A0L7KZ77 A0A067R4Q3 A0A3B4UI57 A0A3B4YN70 A0A3Q3A5T0 A0A3P9AF43 A0A1B6DRI5 A0A3B4ZZD4 H2T4E8 A0A3P8SH69 A0A3Q3MNI8 A0A2I0M6D2 A0A3Q1J506 H0YPF6 A0A3P8YU47 A0A2C9GPL6 K7GFI5 A0A218UMB5 H9FW81 A0A182RE55 G7NWQ8 A0A2K5V853 A0A182KNN6 A0A182JVZ3 A0A2K5K6S7 A0A3P8WCD6 G7MG66 A0A2K6PID3 A0A2K5LDA7 U3JS33 A0A3P8W6Y7 H9ZAH6 K7D9U0 J3SEG9 A0A182N1Y4 A0A182UV55 A0A091IJP4 A0A1S3WVZ1 A0A099YYZ5 A0A182YR49 U3D705 A0A091VIB7 U3F0M8 Q5H8A4-2 T1DMX4 K7BYG4 A0A2J8SSF0 A0A093PJP2 H9G5I3 A0A182J5P2 A0A091FDR4 K7GFH3 A0A091VDW9 A0A0Q3PVM7 Q5H8A4 F6SKB9 V9KC18 G1QJB2 A0A2I0UNF3 A0A2K6BI25 A0A0A0ABJ5 A0A091J0Z8 A0A182QI15 A0A2J8SSG2 W4Y790 F7HKG0 A0A2K6JU75 A0A1S3WWB8 A0A2K6PIM3 A0A0D9RRK0 A0A2K5E4N3 A0A2K5QQ78 A0A1U7QN93 M0RDM7 M3W965 A0A3Q7QB77 A0A1L8HN52 A0A2R9A5W5 H2QP26 A0A2Y9G5I0 E7EWV1 A0A2K6ETQ2 H0WI69 L8IWH2 A0A182TYI6 Q8NCI4 A0A3Q0DM45 F6VAS9 A0A3Q7VV01 A0A3Q0G8V4 A0A3Q0G5C2
A0A2H1WSG9 A0A212EW94 A0A0L7KZ77 A0A067R4Q3 A0A3B4UI57 A0A3B4YN70 A0A3Q3A5T0 A0A3P9AF43 A0A1B6DRI5 A0A3B4ZZD4 H2T4E8 A0A3P8SH69 A0A3Q3MNI8 A0A2I0M6D2 A0A3Q1J506 H0YPF6 A0A3P8YU47 A0A2C9GPL6 K7GFI5 A0A218UMB5 H9FW81 A0A182RE55 G7NWQ8 A0A2K5V853 A0A182KNN6 A0A182JVZ3 A0A2K5K6S7 A0A3P8WCD6 G7MG66 A0A2K6PID3 A0A2K5LDA7 U3JS33 A0A3P8W6Y7 H9ZAH6 K7D9U0 J3SEG9 A0A182N1Y4 A0A182UV55 A0A091IJP4 A0A1S3WVZ1 A0A099YYZ5 A0A182YR49 U3D705 A0A091VIB7 U3F0M8 Q5H8A4-2 T1DMX4 K7BYG4 A0A2J8SSF0 A0A093PJP2 H9G5I3 A0A182J5P2 A0A091FDR4 K7GFH3 A0A091VDW9 A0A0Q3PVM7 Q5H8A4 F6SKB9 V9KC18 G1QJB2 A0A2I0UNF3 A0A2K6BI25 A0A0A0ABJ5 A0A091J0Z8 A0A182QI15 A0A2J8SSG2 W4Y790 F7HKG0 A0A2K6JU75 A0A1S3WWB8 A0A2K6PIM3 A0A0D9RRK0 A0A2K5E4N3 A0A2K5QQ78 A0A1U7QN93 M0RDM7 M3W965 A0A3Q7QB77 A0A1L8HN52 A0A2R9A5W5 H2QP26 A0A2Y9G5I0 E7EWV1 A0A2K6ETQ2 H0WI69 L8IWH2 A0A182TYI6 Q8NCI4 A0A3Q0DM45 F6VAS9 A0A3Q7VV01 A0A3Q0G8V4 A0A3Q0G5C2
EC Number
2.-.-.-
Pubmed
26354079
19121390
22118469
26227816
24845553
25069045
+ More
21551351 23371554 20360741 17381049 25319552 22002653 20966253 24487278 25362486 23025625 25244985 25243066 15632136 12975309 16303743 14702039 15815621 15489334 26996948 23758969 25727380 21881562 17495919 24402279 17431167 15057822 17975172 27762356 22722832 16136131 22751099 20431018
21551351 23371554 20360741 17381049 25319552 22002653 20966253 24487278 25362486 23025625 25244985 25243066 15632136 12975309 16303743 14702039 15815621 15489334 26996948 23758969 25727380 21881562 17495919 24402279 17431167 15057822 17975172 27762356 22722832 16136131 22751099 20431018
EMBL
ODYU01000850
SOQ36237.1
RSAL01000096
RVE47703.1
NWSH01001992
PCG69498.1
+ More
KQ459185 KPJ03296.1 KQ461073 KPJ09604.1 BABH01021572 BABH01021573 BABH01021574 BABH01021575 ODYU01010708 SOQ55988.1 AGBW02012080 OWR45714.1 JTDY01004149 KOB68527.1 KK852981 KDR12958.1 GEDC01009000 JAS28298.1 AKCR02000034 PKK25239.1 ABQF01041685 ABQF01041686 ABQF01041687 ABQF01041688 ABQF01041689 ABQF01041690 ABQF01041691 ABQF01041692 APCN01004657 AGCU01006575 AGCU01006576 AGCU01006577 AGCU01006578 AGCU01006579 AGCU01006580 AGCU01006581 MUZQ01000235 OWK54570.1 JU335137 JV047461 AFE78890.1 AFI37532.1 CM001276 EHH49448.1 AQIA01053376 AQIA01053377 AQIA01053378 AQIA01053379 AQIA01053380 AQIA01053381 CM001253 EHH14205.1 AGTO01011095 JU476138 AFH32942.1 GABC01005165 GABF01003032 GABD01008606 GABE01001696 NBAG03000389 JAA06173.1 JAA19113.1 JAA24494.1 JAA43043.1 PNI30843.1 JU174700 GBEX01001430 AFJ50226.1 JAI13130.1 KL218793 KFP08492.1 KL887216 KGL74656.1 GAMR01006010 JAB27922.1 KL411079 KFR03147.1 GAMT01003635 GAMS01009608 GAMP01001953 JAB08226.1 JAB13528.1 JAB50802.1 AB162713 AY358538 AK074815 AK000272 AK027465 AK097244 AK296507 AC092574 AC116565 BC000937 BC001249 BC110878 GAAZ01001017 GBKC01001510 JAA96926.1 JAG44560.1 GABC01005166 GABF01003031 GABD01008607 GABE01001697 JAA06172.1 JAA19114.1 JAA24493.1 JAA43042.1 PNI30842.1 NDHI03003552 PNJ23708.1 KL225457 KFW72402.1 AAWZ02013353 AAWZ02013354 KK718913 KFO59800.1 KK733874 KFR00935.1 LMAW01002545 KQK80180.1 JW862803 AFO95320.1 ADFV01125059 ADFV01125060 ADFV01125061 KZ505675 PKU47557.1 KL871357 KGL91909.1 KK501335 KFP14282.1 AXCN02000977 PNJ23707.1 AAGJ04096092 JSUE03032701 JSUE03032702 JSUE03032703 AQIB01147730 AQIB01147731 AQIB01147732 AABR07014132 AC103314 AANG04001605 CM004467 OCT97514.1 AJFE02035480 AACZ04061257 PNI30844.1 AAQR03169398 AAQR03169399 AAQR03169400 AAQR03169401 JH880544 ELR60801.1 AK074715 BAC11157.1 AAMC01111117 AAMC01111118 AAMC01111119 AAMC01111120
KQ459185 KPJ03296.1 KQ461073 KPJ09604.1 BABH01021572 BABH01021573 BABH01021574 BABH01021575 ODYU01010708 SOQ55988.1 AGBW02012080 OWR45714.1 JTDY01004149 KOB68527.1 KK852981 KDR12958.1 GEDC01009000 JAS28298.1 AKCR02000034 PKK25239.1 ABQF01041685 ABQF01041686 ABQF01041687 ABQF01041688 ABQF01041689 ABQF01041690 ABQF01041691 ABQF01041692 APCN01004657 AGCU01006575 AGCU01006576 AGCU01006577 AGCU01006578 AGCU01006579 AGCU01006580 AGCU01006581 MUZQ01000235 OWK54570.1 JU335137 JV047461 AFE78890.1 AFI37532.1 CM001276 EHH49448.1 AQIA01053376 AQIA01053377 AQIA01053378 AQIA01053379 AQIA01053380 AQIA01053381 CM001253 EHH14205.1 AGTO01011095 JU476138 AFH32942.1 GABC01005165 GABF01003032 GABD01008606 GABE01001696 NBAG03000389 JAA06173.1 JAA19113.1 JAA24494.1 JAA43043.1 PNI30843.1 JU174700 GBEX01001430 AFJ50226.1 JAI13130.1 KL218793 KFP08492.1 KL887216 KGL74656.1 GAMR01006010 JAB27922.1 KL411079 KFR03147.1 GAMT01003635 GAMS01009608 GAMP01001953 JAB08226.1 JAB13528.1 JAB50802.1 AB162713 AY358538 AK074815 AK000272 AK027465 AK097244 AK296507 AC092574 AC116565 BC000937 BC001249 BC110878 GAAZ01001017 GBKC01001510 JAA96926.1 JAG44560.1 GABC01005166 GABF01003031 GABD01008607 GABE01001697 JAA06172.1 JAA19114.1 JAA24493.1 JAA43042.1 PNI30842.1 NDHI03003552 PNJ23708.1 KL225457 KFW72402.1 AAWZ02013353 AAWZ02013354 KK718913 KFO59800.1 KK733874 KFR00935.1 LMAW01002545 KQK80180.1 JW862803 AFO95320.1 ADFV01125059 ADFV01125060 ADFV01125061 KZ505675 PKU47557.1 KL871357 KGL91909.1 KK501335 KFP14282.1 AXCN02000977 PNJ23707.1 AAGJ04096092 JSUE03032701 JSUE03032702 JSUE03032703 AQIB01147730 AQIB01147731 AQIB01147732 AABR07014132 AC103314 AANG04001605 CM004467 OCT97514.1 AJFE02035480 AACZ04061257 PNI30844.1 AAQR03169398 AAQR03169399 AAQR03169400 AAQR03169401 JH880544 ELR60801.1 AK074715 BAC11157.1 AAMC01111117 AAMC01111118 AAMC01111119 AAMC01111120
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000005204
UP000007151
+ More
UP000037510 UP000027135 UP000261420 UP000261360 UP000264800 UP000265140 UP000261400 UP000005226 UP000265080 UP000261640 UP000053872 UP000265040 UP000007754 UP000075840 UP000007267 UP000197619 UP000075900 UP000009130 UP000233100 UP000075882 UP000075881 UP000233080 UP000265120 UP000233200 UP000233060 UP000016665 UP000075884 UP000075903 UP000054308 UP000079721 UP000053641 UP000076408 UP000053283 UP000008225 UP000005640 UP000054081 UP000001646 UP000075880 UP000052976 UP000053605 UP000051836 UP000002280 UP000001073 UP000233120 UP000053858 UP000053119 UP000075886 UP000007110 UP000006718 UP000233180 UP000029965 UP000233020 UP000233040 UP000189706 UP000002494 UP000011712 UP000286641 UP000186698 UP000240080 UP000002277 UP000248481 UP000233160 UP000005225 UP000075902 UP000189704 UP000008143 UP000286642 UP000189705
UP000037510 UP000027135 UP000261420 UP000261360 UP000264800 UP000265140 UP000261400 UP000005226 UP000265080 UP000261640 UP000053872 UP000265040 UP000007754 UP000075840 UP000007267 UP000197619 UP000075900 UP000009130 UP000233100 UP000075882 UP000075881 UP000233080 UP000265120 UP000233200 UP000233060 UP000016665 UP000075884 UP000075903 UP000054308 UP000079721 UP000053641 UP000076408 UP000053283 UP000008225 UP000005640 UP000054081 UP000001646 UP000075880 UP000052976 UP000053605 UP000051836 UP000002280 UP000001073 UP000233120 UP000053858 UP000053119 UP000075886 UP000007110 UP000006718 UP000233180 UP000029965 UP000233020 UP000233040 UP000189706 UP000002494 UP000011712 UP000286641 UP000186698 UP000240080 UP000002277 UP000248481 UP000233160 UP000005225 UP000075902 UP000189704 UP000008143 UP000286642 UP000189705
Pfam
PF01663 Phosphodiest
Interpro
SUPFAM
SSF53649
SSF53649
Gene 3D
ProteinModelPortal
A0A2H1V5Z2
A0A3S2LZI0
A0A2A4JCS8
A0A194QIL6
A0A194QWC4
H9J3D0
+ More
A0A2H1WSG9 A0A212EW94 A0A0L7KZ77 A0A067R4Q3 A0A3B4UI57 A0A3B4YN70 A0A3Q3A5T0 A0A3P9AF43 A0A1B6DRI5 A0A3B4ZZD4 H2T4E8 A0A3P8SH69 A0A3Q3MNI8 A0A2I0M6D2 A0A3Q1J506 H0YPF6 A0A3P8YU47 A0A2C9GPL6 K7GFI5 A0A218UMB5 H9FW81 A0A182RE55 G7NWQ8 A0A2K5V853 A0A182KNN6 A0A182JVZ3 A0A2K5K6S7 A0A3P8WCD6 G7MG66 A0A2K6PID3 A0A2K5LDA7 U3JS33 A0A3P8W6Y7 H9ZAH6 K7D9U0 J3SEG9 A0A182N1Y4 A0A182UV55 A0A091IJP4 A0A1S3WVZ1 A0A099YYZ5 A0A182YR49 U3D705 A0A091VIB7 U3F0M8 Q5H8A4-2 T1DMX4 K7BYG4 A0A2J8SSF0 A0A093PJP2 H9G5I3 A0A182J5P2 A0A091FDR4 K7GFH3 A0A091VDW9 A0A0Q3PVM7 Q5H8A4 F6SKB9 V9KC18 G1QJB2 A0A2I0UNF3 A0A2K6BI25 A0A0A0ABJ5 A0A091J0Z8 A0A182QI15 A0A2J8SSG2 W4Y790 F7HKG0 A0A2K6JU75 A0A1S3WWB8 A0A2K6PIM3 A0A0D9RRK0 A0A2K5E4N3 A0A2K5QQ78 A0A1U7QN93 M0RDM7 M3W965 A0A3Q7QB77 A0A1L8HN52 A0A2R9A5W5 H2QP26 A0A2Y9G5I0 E7EWV1 A0A2K6ETQ2 H0WI69 L8IWH2 A0A182TYI6 Q8NCI4 A0A3Q0DM45 F6VAS9 A0A3Q7VV01 A0A3Q0G8V4 A0A3Q0G5C2
A0A2H1WSG9 A0A212EW94 A0A0L7KZ77 A0A067R4Q3 A0A3B4UI57 A0A3B4YN70 A0A3Q3A5T0 A0A3P9AF43 A0A1B6DRI5 A0A3B4ZZD4 H2T4E8 A0A3P8SH69 A0A3Q3MNI8 A0A2I0M6D2 A0A3Q1J506 H0YPF6 A0A3P8YU47 A0A2C9GPL6 K7GFI5 A0A218UMB5 H9FW81 A0A182RE55 G7NWQ8 A0A2K5V853 A0A182KNN6 A0A182JVZ3 A0A2K5K6S7 A0A3P8WCD6 G7MG66 A0A2K6PID3 A0A2K5LDA7 U3JS33 A0A3P8W6Y7 H9ZAH6 K7D9U0 J3SEG9 A0A182N1Y4 A0A182UV55 A0A091IJP4 A0A1S3WVZ1 A0A099YYZ5 A0A182YR49 U3D705 A0A091VIB7 U3F0M8 Q5H8A4-2 T1DMX4 K7BYG4 A0A2J8SSF0 A0A093PJP2 H9G5I3 A0A182J5P2 A0A091FDR4 K7GFH3 A0A091VDW9 A0A0Q3PVM7 Q5H8A4 F6SKB9 V9KC18 G1QJB2 A0A2I0UNF3 A0A2K6BI25 A0A0A0ABJ5 A0A091J0Z8 A0A182QI15 A0A2J8SSG2 W4Y790 F7HKG0 A0A2K6JU75 A0A1S3WWB8 A0A2K6PIM3 A0A0D9RRK0 A0A2K5E4N3 A0A2K5QQ78 A0A1U7QN93 M0RDM7 M3W965 A0A3Q7QB77 A0A1L8HN52 A0A2R9A5W5 H2QP26 A0A2Y9G5I0 E7EWV1 A0A2K6ETQ2 H0WI69 L8IWH2 A0A182TYI6 Q8NCI4 A0A3Q0DM45 F6VAS9 A0A3Q7VV01 A0A3Q0G8V4 A0A3Q0G5C2
PDB
1O99
E-value=0.00113378,
Score=104
Ontologies
PATHWAY
GO
PANTHER
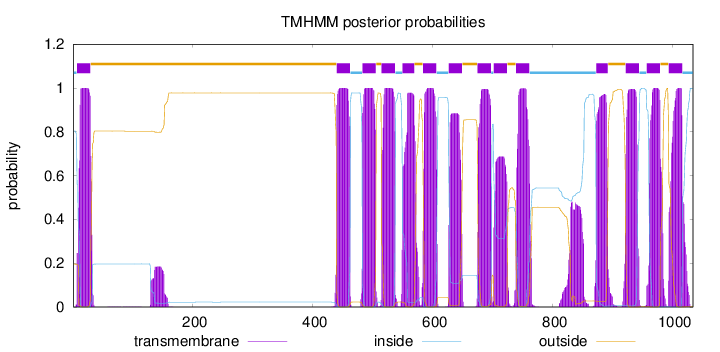
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
1034
Number of predicted TMHs:
14
Exp number of AAs in TMHs:
312.12575
Exp number, first 60 AAs:
21.83914
Total prob of N-in:
0.80374
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 439
TMhelix
440 - 462
inside
463 - 482
TMhelix
483 - 505
outside
506 - 514
TMhelix
515 - 537
inside
538 - 549
TMhelix
550 - 569
outside
570 - 583
TMhelix
584 - 606
inside
607 - 626
TMhelix
627 - 649
outside
650 - 674
TMhelix
675 - 697
inside
698 - 701
TMhelix
702 - 724
outside
725 - 738
TMhelix
739 - 761
inside
762 - 872
TMhelix
873 - 892
outside
893 - 921
TMhelix
922 - 944
inside
945 - 956
TMhelix
957 - 979
outside
980 - 993
TMhelix
994 - 1016
inside
1017 - 1034
Population Genetic Test Statistics
Pi
195.327648
Theta
137.763776
Tajima's D
0.434444
CLR
1.422444
CSRT
0.489725513724314
Interpretation
Uncertain
