Pre Gene Modal
BGIBMGA004102
Annotation
PREDICTED:_peptidyl-prolyl_cis-trans_isomerase_FKBP8_isoform_X2_[Papilio_machaon]
Full name
Peptidylprolyl isomerase
Location in the cell
Cytoplasmic Reliability : 2.624
Sequence
CDS
ATGAGTGAAGAAAGCAAAGCTTTGACGGACAAGTCTGAAAGCAGCTCTTTCGAAGACTTAGCGGCTGCGGCTGCTGAGGAGATCGAGGAAGCGAAACTTGCCGAAGCTGCCGCAGCAGAGAAGGAAAAATCCCCAGAAGTAAAGAATGACGAATGGCAAGATGTTCTTGGGTCCGGAGCTCTGCTTAAAAAGATATTGAAACAAGGGGATGAAATATCTGGATCCAGACCACAAAGAGGGGACATATGCAGAATCAGCTATGAACTCAAAATAAAAGACAGTAACAACATTGTGGAGAAAAGAGATCAGATAAAAATATATTTAGGAGACAATGAGGTATTGCAAGGTTTGGACCTAGCATTAACTTTAATGTACAGAGGTGAGGAATGTATTTTGCAACTTGCTCCAAGATTTGCCTATGGAGAAATGGGACTTAAACCAGGTGAGAGCCTTGGATTAGTCGGACAGTGTGATGAACCCAAGTATAAAGGACCTATTATTGGACCAGACACATGGCTTGAAGCCAAATTAGTGCTGCATGATTGGTCAGAAGAACCTGAACACGATGTATTGTCAATAGCGGAAAAAATGGAAATCGGAATTCGTCGTAGAGCCCGCGGTAACTGGTGGTACGGTCGTGATGAACCACAACTAGCGGTGCAGCTATATCGGAGAGCTCTCGACATTCTGGACGAAAGCGAAGGAGGGATCACTGATCCCACGCCGTCTGGGGAACTCACATTCGCTAACCAGGCACTGAAAGAGTTGCTGGATGAACGACTCAGGGTTCATAACAACATGGCTGCTGCTCAACTCAAGGCCGGAGCTTATGAAGCTGCTTTACAGGCCGTAACCAGAGTACTTTCATGTCAACCGAAAAACGCTAAAGCTCTTTATCGCAAATCACGCATTCTATCTGCGATGGGACGGAACCCTGAAGCCCTGGAGGCAGCTAAAGCAGCGGCGGCCGCAGCCCCCGAGGATACTGGTGCTAAAAAAGAATTACAACGATGCGAACAAAGAGCTTCTAGGGATCGTTCTGTGGAAAGGAAACTGGCTAAACGTATGCTGGGCACTACTGACGTGTCGGCGAAAAGCGATGCACCCTCTCCCGATAAGAAACCTTCCAGGTCTAAGATCCTAGTGTGGGGCTCGCTTCTGCTCAGCTTGTTGGTGGGTGTCGCCAGCGTCCTCGTATACAGATATAAAATGCAGGCGCACTGA
Protein
MSEESKALTDKSESSSFEDLAAAAAEEIEEAKLAEAAAAEKEKSPEVKNDEWQDVLGSGALLKKILKQGDEISGSRPQRGDICRISYELKIKDSNNIVEKRDQIKIYLGDNEVLQGLDLALTLMYRGEECILQLAPRFAYGEMGLKPGESLGLVGQCDEPKYKGPIIGPDTWLEAKLVLHDWSEEPEHDVLSIAEKMEIGIRRRARGNWWYGRDEPQLAVQLYRRALDILDESEGGITDPTPSGELTFANQALKELLDERLRVHNNMAAAQLKAGAYEAALQAVTRVLSCQPKNAKALYRKSRILSAMGRNPEALEAAKAAAAAAPEDTGAKKELQRCEQRASRDRSVERKLAKRMLGTTDVSAKSDAPSPDKKPSRSKILVWGSLLLSLLVGVASVLVYRYKMQAH
Summary
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Uniprot
H9J3L5
A0A194QCV5
A0A3S2LJ00
A0A2A4JBM8
A0A2A4JCM2
A0A2H1V5V9
+ More
A0A212EW45 A0A1E1WQF4 A0A194R188 A0A0L7KZM2 A0A1B6CIU3 A0A067R311 A0A023EQ01 A0A2J7RJX8 U5EVG1 A0A224XIY8 A0A232FL82 A0A1S4FVM5 Q16MK5 A0A069DT63 A0A1S4FVT6 K7JJZ4 E2C4I6 Q16MK6 A0A0P4VRE9 A0A023F1H6 A0A1Y1KQQ1 A0A1Y1KRE4 D6WQV5 A0A2R7X2F8 T1HDK6 A0A0K8TPX4 A0A161ML76 A0A1W4WGY4 A0A1W4W6J4 A0A310SUB3 A0A1J1IEC5 A0A088A5M5 A0A1Q3F7K8 A0A1Q3F7L9 A0A1Q3F7H5 A0A182UVC9 A0A2A3E531 A0A182X4J8 Q7PI62 A0A182TWS1 A0A348G6B6 A0A182I4J5 B0WD01 A0A154PUR8 A0A182R965 A0A0A9YRZ0 W5J8E1 A0A0J7KMC0 A0A2M4BPR3 A0A2M4AAM2 A0A182PMV5 A0A1B0GJM0 A0A182JU29 A0A0L7R2M9 A0A2M3Z3F8 A0A182FGF4 A0A1L8DTW8 A0A1B6JU69 A0A2M4BQM1 A0A2M4A951 A0A0K8TJ23 A0A1I8MHG7 A0A1I8PY78 J3JVI7 A0A2M3Z374 A0A1L8DTN4 A0A182QGN6 A0A182NI90 A0A2M3ZGY7 A0A182LUU9 A0A1L8ECL2 A0A1L8DTP9 T1PC14 A0A182WBQ0 A0A182IXF5 A0A1L8ECM2 A0A1B6FC90 E0VH54 T1DHC7 A0A182Y2J2 A0A0K8W9Z2 A0A0P5HR46 A0A0P6DA46 A0A0N8CKH4 A0A0P5LA10 W8AHP7 A0A0P5RR79 A0A0P5LEB9 A0A0N8BJ23
A0A212EW45 A0A1E1WQF4 A0A194R188 A0A0L7KZM2 A0A1B6CIU3 A0A067R311 A0A023EQ01 A0A2J7RJX8 U5EVG1 A0A224XIY8 A0A232FL82 A0A1S4FVM5 Q16MK5 A0A069DT63 A0A1S4FVT6 K7JJZ4 E2C4I6 Q16MK6 A0A0P4VRE9 A0A023F1H6 A0A1Y1KQQ1 A0A1Y1KRE4 D6WQV5 A0A2R7X2F8 T1HDK6 A0A0K8TPX4 A0A161ML76 A0A1W4WGY4 A0A1W4W6J4 A0A310SUB3 A0A1J1IEC5 A0A088A5M5 A0A1Q3F7K8 A0A1Q3F7L9 A0A1Q3F7H5 A0A182UVC9 A0A2A3E531 A0A182X4J8 Q7PI62 A0A182TWS1 A0A348G6B6 A0A182I4J5 B0WD01 A0A154PUR8 A0A182R965 A0A0A9YRZ0 W5J8E1 A0A0J7KMC0 A0A2M4BPR3 A0A2M4AAM2 A0A182PMV5 A0A1B0GJM0 A0A182JU29 A0A0L7R2M9 A0A2M3Z3F8 A0A182FGF4 A0A1L8DTW8 A0A1B6JU69 A0A2M4BQM1 A0A2M4A951 A0A0K8TJ23 A0A1I8MHG7 A0A1I8PY78 J3JVI7 A0A2M3Z374 A0A1L8DTN4 A0A182QGN6 A0A182NI90 A0A2M3ZGY7 A0A182LUU9 A0A1L8ECL2 A0A1L8DTP9 T1PC14 A0A182WBQ0 A0A182IXF5 A0A1L8ECM2 A0A1B6FC90 E0VH54 T1DHC7 A0A182Y2J2 A0A0K8W9Z2 A0A0P5HR46 A0A0P6DA46 A0A0N8CKH4 A0A0P5LA10 W8AHP7 A0A0P5RR79 A0A0P5LEB9 A0A0N8BJ23
EC Number
5.2.1.8
Pubmed
EMBL
BABH01021572
KQ459185
KPJ03297.1
RSAL01000096
RVE47704.1
NWSH01001992
+ More
PCG69497.1 PCG69496.1 ODYU01000850 SOQ36235.1 AGBW02012080 OWR45712.1 GDQN01001846 GDQN01000382 JAT89208.1 JAT90672.1 KQ461073 KPJ09606.1 JTDY01004149 KOB68526.1 GEDC01028241 GEDC01023902 GEDC01020126 GEDC01001016 JAS09057.1 JAS13396.1 JAS17172.1 JAS36282.1 KK853041 KDR12172.1 JXUM01000446 GAPW01001971 JAC11627.1 NEVH01002983 PNF41142.1 GANO01003420 JAB56451.1 GFTR01005412 JAW11014.1 NNAY01000089 OXU31097.1 CH477859 EAT35561.1 GBGD01001978 JAC86911.1 GL452471 EFN77170.1 EAT35560.1 GDKW01002985 JAI53610.1 GBBI01003846 JAC14866.1 GEZM01080890 JAV62005.1 GEZM01080889 JAV62006.1 KQ971351 EFA06496.1 KK856469 PTY25841.1 ACPB03003322 ACPB03003323 GDAI01001422 JAI16181.1 GEMB01002508 JAS00678.1 KQ760116 OAD61952.1 CVRI01000047 CRK98122.1 GFDL01011518 JAV23527.1 GFDL01011490 JAV23555.1 GFDL01011525 JAV23520.1 KZ288400 PBC26279.1 AAAB01008960 EAA44266.3 FX986032 BBF97989.1 APCN01002316 DS231892 EDS44076.1 KQ435178 KZC14950.1 GBHO01025269 GBHO01012453 GBHO01009736 GBHO01009735 GDHC01019141 GDHC01011628 JAG18335.1 JAG31151.1 JAG33868.1 JAG33869.1 JAP99487.1 JAQ07001.1 ADMH02002104 ETN59059.1 LBMM01005605 KMQ91381.1 GGFJ01005926 MBW55067.1 GGFK01004515 MBW37836.1 AJWK01021981 AJWK01021982 AJWK01021983 KQ414665 KOC65132.1 GGFM01002227 MBW22978.1 GFDF01004250 JAV09834.1 GECU01004965 JAT02742.1 GGFJ01005927 MBW55068.1 GGFK01004005 MBW37326.1 GBRD01000693 GBRD01000692 GBRD01000691 GBRD01000690 JAG65130.1 BT127255 AEE62217.1 GGFM01002228 MBW22979.1 GFDF01004252 JAV09832.1 AXCN02000877 GGFM01006959 MBW27710.1 AXCM01001383 GFDG01002329 JAV16470.1 GFDF01004251 JAV09833.1 KA646311 AFP60940.1 GFDG01002328 JAV16471.1 GECZ01021944 JAS47825.1 DS235161 EEB12710.1 GAMD01002426 JAA99164.1 GDHF01004338 JAI47976.1 GDIQ01229903 JAK21822.1 GDIP01132057 GDIQ01082007 JAL71657.1 JAN12730.1 GDIP01122855 JAL80859.1 GDIQ01172873 JAK78852.1 GAMC01018560 JAB87995.1 GDIQ01097153 JAL54573.1 GDIQ01175274 JAK76451.1 GDIP01170451 GDIQ01172875 JAJ52951.1 JAK78850.1
PCG69497.1 PCG69496.1 ODYU01000850 SOQ36235.1 AGBW02012080 OWR45712.1 GDQN01001846 GDQN01000382 JAT89208.1 JAT90672.1 KQ461073 KPJ09606.1 JTDY01004149 KOB68526.1 GEDC01028241 GEDC01023902 GEDC01020126 GEDC01001016 JAS09057.1 JAS13396.1 JAS17172.1 JAS36282.1 KK853041 KDR12172.1 JXUM01000446 GAPW01001971 JAC11627.1 NEVH01002983 PNF41142.1 GANO01003420 JAB56451.1 GFTR01005412 JAW11014.1 NNAY01000089 OXU31097.1 CH477859 EAT35561.1 GBGD01001978 JAC86911.1 GL452471 EFN77170.1 EAT35560.1 GDKW01002985 JAI53610.1 GBBI01003846 JAC14866.1 GEZM01080890 JAV62005.1 GEZM01080889 JAV62006.1 KQ971351 EFA06496.1 KK856469 PTY25841.1 ACPB03003322 ACPB03003323 GDAI01001422 JAI16181.1 GEMB01002508 JAS00678.1 KQ760116 OAD61952.1 CVRI01000047 CRK98122.1 GFDL01011518 JAV23527.1 GFDL01011490 JAV23555.1 GFDL01011525 JAV23520.1 KZ288400 PBC26279.1 AAAB01008960 EAA44266.3 FX986032 BBF97989.1 APCN01002316 DS231892 EDS44076.1 KQ435178 KZC14950.1 GBHO01025269 GBHO01012453 GBHO01009736 GBHO01009735 GDHC01019141 GDHC01011628 JAG18335.1 JAG31151.1 JAG33868.1 JAG33869.1 JAP99487.1 JAQ07001.1 ADMH02002104 ETN59059.1 LBMM01005605 KMQ91381.1 GGFJ01005926 MBW55067.1 GGFK01004515 MBW37836.1 AJWK01021981 AJWK01021982 AJWK01021983 KQ414665 KOC65132.1 GGFM01002227 MBW22978.1 GFDF01004250 JAV09834.1 GECU01004965 JAT02742.1 GGFJ01005927 MBW55068.1 GGFK01004005 MBW37326.1 GBRD01000693 GBRD01000692 GBRD01000691 GBRD01000690 JAG65130.1 BT127255 AEE62217.1 GGFM01002228 MBW22979.1 GFDF01004252 JAV09832.1 AXCN02000877 GGFM01006959 MBW27710.1 AXCM01001383 GFDG01002329 JAV16470.1 GFDF01004251 JAV09833.1 KA646311 AFP60940.1 GFDG01002328 JAV16471.1 GECZ01021944 JAS47825.1 DS235161 EEB12710.1 GAMD01002426 JAA99164.1 GDHF01004338 JAI47976.1 GDIQ01229903 JAK21822.1 GDIP01132057 GDIQ01082007 JAL71657.1 JAN12730.1 GDIP01122855 JAL80859.1 GDIQ01172873 JAK78852.1 GAMC01018560 JAB87995.1 GDIQ01097153 JAL54573.1 GDIQ01175274 JAK76451.1 GDIP01170451 GDIQ01172875 JAJ52951.1 JAK78850.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000218220
UP000007151
UP000053240
+ More
UP000037510 UP000027135 UP000069940 UP000235965 UP000215335 UP000008820 UP000002358 UP000008237 UP000007266 UP000015103 UP000192223 UP000183832 UP000005203 UP000075903 UP000242457 UP000076407 UP000007062 UP000075902 UP000075840 UP000002320 UP000076502 UP000075900 UP000000673 UP000036403 UP000075885 UP000092461 UP000075881 UP000053825 UP000069272 UP000095301 UP000095300 UP000075886 UP000075884 UP000075883 UP000075920 UP000075880 UP000009046 UP000076408
UP000037510 UP000027135 UP000069940 UP000235965 UP000215335 UP000008820 UP000002358 UP000008237 UP000007266 UP000015103 UP000192223 UP000183832 UP000005203 UP000075903 UP000242457 UP000076407 UP000007062 UP000075902 UP000075840 UP000002320 UP000076502 UP000075900 UP000000673 UP000036403 UP000075885 UP000092461 UP000075881 UP000053825 UP000069272 UP000095301 UP000095300 UP000075886 UP000075884 UP000075883 UP000075920 UP000075880 UP000009046 UP000076408
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9J3L5
A0A194QCV5
A0A3S2LJ00
A0A2A4JBM8
A0A2A4JCM2
A0A2H1V5V9
+ More
A0A212EW45 A0A1E1WQF4 A0A194R188 A0A0L7KZM2 A0A1B6CIU3 A0A067R311 A0A023EQ01 A0A2J7RJX8 U5EVG1 A0A224XIY8 A0A232FL82 A0A1S4FVM5 Q16MK5 A0A069DT63 A0A1S4FVT6 K7JJZ4 E2C4I6 Q16MK6 A0A0P4VRE9 A0A023F1H6 A0A1Y1KQQ1 A0A1Y1KRE4 D6WQV5 A0A2R7X2F8 T1HDK6 A0A0K8TPX4 A0A161ML76 A0A1W4WGY4 A0A1W4W6J4 A0A310SUB3 A0A1J1IEC5 A0A088A5M5 A0A1Q3F7K8 A0A1Q3F7L9 A0A1Q3F7H5 A0A182UVC9 A0A2A3E531 A0A182X4J8 Q7PI62 A0A182TWS1 A0A348G6B6 A0A182I4J5 B0WD01 A0A154PUR8 A0A182R965 A0A0A9YRZ0 W5J8E1 A0A0J7KMC0 A0A2M4BPR3 A0A2M4AAM2 A0A182PMV5 A0A1B0GJM0 A0A182JU29 A0A0L7R2M9 A0A2M3Z3F8 A0A182FGF4 A0A1L8DTW8 A0A1B6JU69 A0A2M4BQM1 A0A2M4A951 A0A0K8TJ23 A0A1I8MHG7 A0A1I8PY78 J3JVI7 A0A2M3Z374 A0A1L8DTN4 A0A182QGN6 A0A182NI90 A0A2M3ZGY7 A0A182LUU9 A0A1L8ECL2 A0A1L8DTP9 T1PC14 A0A182WBQ0 A0A182IXF5 A0A1L8ECM2 A0A1B6FC90 E0VH54 T1DHC7 A0A182Y2J2 A0A0K8W9Z2 A0A0P5HR46 A0A0P6DA46 A0A0N8CKH4 A0A0P5LA10 W8AHP7 A0A0P5RR79 A0A0P5LEB9 A0A0N8BJ23
A0A212EW45 A0A1E1WQF4 A0A194R188 A0A0L7KZM2 A0A1B6CIU3 A0A067R311 A0A023EQ01 A0A2J7RJX8 U5EVG1 A0A224XIY8 A0A232FL82 A0A1S4FVM5 Q16MK5 A0A069DT63 A0A1S4FVT6 K7JJZ4 E2C4I6 Q16MK6 A0A0P4VRE9 A0A023F1H6 A0A1Y1KQQ1 A0A1Y1KRE4 D6WQV5 A0A2R7X2F8 T1HDK6 A0A0K8TPX4 A0A161ML76 A0A1W4WGY4 A0A1W4W6J4 A0A310SUB3 A0A1J1IEC5 A0A088A5M5 A0A1Q3F7K8 A0A1Q3F7L9 A0A1Q3F7H5 A0A182UVC9 A0A2A3E531 A0A182X4J8 Q7PI62 A0A182TWS1 A0A348G6B6 A0A182I4J5 B0WD01 A0A154PUR8 A0A182R965 A0A0A9YRZ0 W5J8E1 A0A0J7KMC0 A0A2M4BPR3 A0A2M4AAM2 A0A182PMV5 A0A1B0GJM0 A0A182JU29 A0A0L7R2M9 A0A2M3Z3F8 A0A182FGF4 A0A1L8DTW8 A0A1B6JU69 A0A2M4BQM1 A0A2M4A951 A0A0K8TJ23 A0A1I8MHG7 A0A1I8PY78 J3JVI7 A0A2M3Z374 A0A1L8DTN4 A0A182QGN6 A0A182NI90 A0A2M3ZGY7 A0A182LUU9 A0A1L8ECL2 A0A1L8DTP9 T1PC14 A0A182WBQ0 A0A182IXF5 A0A1L8ECM2 A0A1B6FC90 E0VH54 T1DHC7 A0A182Y2J2 A0A0K8W9Z2 A0A0P5HR46 A0A0P6DA46 A0A0N8CKH4 A0A0P5LA10 W8AHP7 A0A0P5RR79 A0A0P5LEB9 A0A0N8BJ23
PDB
5MGX
E-value=1.02562e-25,
Score=290
Ontologies
GO
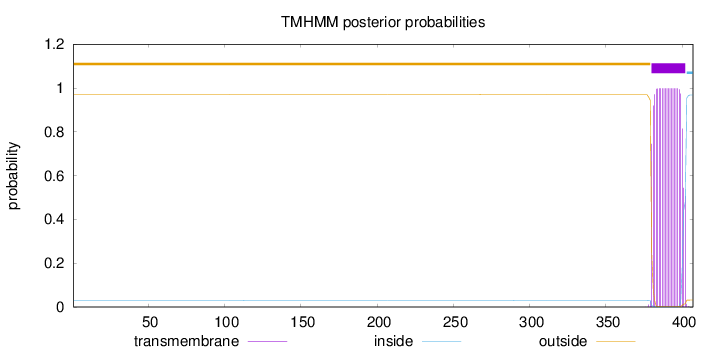
Topology
Length:
407
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.50186
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03026
outside
1 - 379
TMhelix
380 - 402
inside
403 - 407
Population Genetic Test Statistics
Pi
193.988762
Theta
181.987969
Tajima's D
-0.367022
CLR
2.141654
CSRT
0.272386380680966
Interpretation
Uncertain
