Pre Gene Modal
BGIBMGA004018
Annotation
cyclin_dependent_kinase_5_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.935
Sequence
CDS
ATGCAAAAATATGAGAAACTTGAAAAAATCGGTGAAGGAACCTATGGTACGGTATTTAAAGCCAAGAATAAAGAAACACACGAAATCGTAGCGCTCAAACGTGTTAGATTAGACGACGACGATGAAGGAGTACCATCTTCCGCTCTTCGTGAAATATGTCTTCTCAAAGAATTAAAGCATAAAAATATAGTGCGACTTTACGATGTATTGCACAGCGAAAAGAAATTAACCCTAGTGTTTGAACATTGTGATCAAGACTTAAAAAAATACTTCGATAGTTTGAATGGAGAAATAGATCTAGATGTTGTGAAATCATTCATGTATCAATTACTTCGAGGACTAGCTTTTTGTCACAGCCACAACGTTCTCCATAGGGACTTGAAGCCACAGAATCTCTTAATAAACAAGAACGGTGAATTAAAGTTAGCTGATTTTGGTCTGGCCAGAGCTTTTGGCATACCAGTAAAGTGCTATTCCGCTGAAGTTGTAACCTTATGGTACCGGCCTCCTGATGTATTATTTGGTGCTAAGTTGTACACAACAAGCATTGATATGTGGTCTGCTGGATGTATATTTGCAGAACTTGCAAATTCGGGCAGACCGCTATTCCCAGGTTCAGATGTTGATGATCAATTAAAGAGGATTTTTAAGCTTTTAGGTACACCCAATGAGGACACATGGCCTGGAGTAACTCAACTGCCGGACTACAAACCTTTGCCTGTTTATCAGCCAAGTTTAGGCCTCGCCCAAGTTGTACCAAGATTGCCGGCTAGAGGTCGAGATCTACTCGCTCGGCTGCTCACCTGCAATCCAGCTTTAAGAATGCCAGCTGATGATGCAATGGCGCATGCGTATTTCCACGATCTCAATCCGTCAGTCAAGAATGATCGATGTTAA
Protein
MQKYEKLEKIGEGTYGTVFKAKNKETHEIVALKRVRLDDDDEGVPSSALREICLLKELKHKNIVRLYDVLHSEKKLTLVFEHCDQDLKKYFDSLNGEIDLDVVKSFMYQLLRGLAFCHSHNVLHRDLKPQNLLINKNGELKLADFGLARAFGIPVKCYSAEVVTLWYRPPDVLFGAKLYTTSIDMWSAGCIFAELANSGRPLFPGSDVDDQLKRIFKLLGTPNEDTWPGVTQLPDYKPLPVYQPSLGLAQVVPRLPARGRDLLARLLTCNPALRMPADDAMAHAYFHDLNPSVKNDRC
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
A0A2A4JBP9
A0A194QVV7
A0A3S2PCR7
A0A194QCN1
D2IGZ4
A0A2H1V7L0
+ More
A0A212EW69 A0A0L7LKW2 D6WPE8 A0A1W4WYE1 A0A3L8E0L8 A0A1Y1LRB6 E2BU63 K7J0W3 A0A2A3EMF0 A0A087ZW58 A0A067RHK8 A0A026WHB2 F4W6H8 A0A195E4Z0 A0A2J7PH71 A0A158P285 A0A1B6C720 A0A0L7QVC5 T1J8G8 A0A1B6FPD9 A0A1B6KII7 A0A1S3D7F6 A0A1Z5KWL9 A0A087UL74 U4U6W6 T1DCI9 A0A0A9YMH8 L7MEV1 A0A154P546 A0A0K8R794 A0A224YKK2 A0A131YW97 A0A023F5U0 W0G1U1 N6TUL8 A0A2R7W298 A0A023GKE3 A0A226EDC4 A0A023FHB4 E9H5F3 A0A195D4C1 A0A1D2MGB3 A0A0P5J3W9 A0A0P5FHT3 T1JXF9 Q16Y76 A0A182GVY2 A0A1V9Y000 A0A084WJP5 A0A0L8FZI1 A0A1Q3FAB3 J9JP19 B0VZB2 A0A182J8Q3 A0A2M4CSX8 A0A0P5QQZ8 A0A182FHV6 A0A0P5ET06 A0A2H8TII6 A0A182MB04 A0A182WQB3 A0A2S2Q8T2 A0A2M4ATJ1 A0A1S3JI38 A0A182U3P4 A0A1B0D3W9 A0A182YSP5 A0A182QG78 A0A182V432 A0A182NJR3 A0A182JXS9 A0A182L359 A0A182XML7 Q7Q6L9 A0A182IG84 A0A132AHX3 A2I478 D4FUN3 R7UIH3 W5J2U2 T1FZB4 A0A1E1XI53 C1BT30 A0A0K8TLT7 V5HW58 A0A0M3K4Q6 A0A0P5T8L2 K1RBB0 A0A0N8BSY9 A0A0N7ZQ80 F1L117 A0A293M2Z1 A0A2P2I2G9 A0A0J7L331
A0A212EW69 A0A0L7LKW2 D6WPE8 A0A1W4WYE1 A0A3L8E0L8 A0A1Y1LRB6 E2BU63 K7J0W3 A0A2A3EMF0 A0A087ZW58 A0A067RHK8 A0A026WHB2 F4W6H8 A0A195E4Z0 A0A2J7PH71 A0A158P285 A0A1B6C720 A0A0L7QVC5 T1J8G8 A0A1B6FPD9 A0A1B6KII7 A0A1S3D7F6 A0A1Z5KWL9 A0A087UL74 U4U6W6 T1DCI9 A0A0A9YMH8 L7MEV1 A0A154P546 A0A0K8R794 A0A224YKK2 A0A131YW97 A0A023F5U0 W0G1U1 N6TUL8 A0A2R7W298 A0A023GKE3 A0A226EDC4 A0A023FHB4 E9H5F3 A0A195D4C1 A0A1D2MGB3 A0A0P5J3W9 A0A0P5FHT3 T1JXF9 Q16Y76 A0A182GVY2 A0A1V9Y000 A0A084WJP5 A0A0L8FZI1 A0A1Q3FAB3 J9JP19 B0VZB2 A0A182J8Q3 A0A2M4CSX8 A0A0P5QQZ8 A0A182FHV6 A0A0P5ET06 A0A2H8TII6 A0A182MB04 A0A182WQB3 A0A2S2Q8T2 A0A2M4ATJ1 A0A1S3JI38 A0A182U3P4 A0A1B0D3W9 A0A182YSP5 A0A182QG78 A0A182V432 A0A182NJR3 A0A182JXS9 A0A182L359 A0A182XML7 Q7Q6L9 A0A182IG84 A0A132AHX3 A2I478 D4FUN3 R7UIH3 W5J2U2 T1FZB4 A0A1E1XI53 C1BT30 A0A0K8TLT7 V5HW58 A0A0M3K4Q6 A0A0P5T8L2 K1RBB0 A0A0N8BSY9 A0A0N7ZQ80 F1L117 A0A293M2Z1 A0A2P2I2G9 A0A0J7L331
Pubmed
26354079
19121390
22118469
26227816
18362917
19820115
+ More
30249741 28004739 20798317 20075255 24845553 24508170 21719571 21347285 28528879 23537049 25401762 26823975 25576852 28797301 26830274 25474469 24146826 21292972 27289101 17510324 26483478 28327890 24438588 25244985 20966253 12364791 14747013 17210077 26555130 23254933 20920257 23761445 28503490 26369729 25765539 22992520 21685128
30249741 28004739 20798317 20075255 24845553 24508170 21719571 21347285 28528879 23537049 25401762 26823975 25576852 28797301 26830274 25474469 24146826 21292972 27289101 17510324 26483478 28327890 24438588 25244985 20966253 12364791 14747013 17210077 26555130 23254933 20920257 23761445 28503490 26369729 25765539 22992520 21685128
EMBL
NWSH01001992
PCG69495.1
KQ461073
KPJ09607.1
RSAL01000096
RVE47706.1
+ More
KQ459185 KPJ03298.1 BABH01021571 GQ250576 ACT83401.1 ODYU01000850 SOQ36234.1 AGBW02012080 OWR45711.1 JTDY01000700 KOB76173.1 KQ971354 EFA06801.2 QOIP01000002 RLU26207.1 GEZM01054547 JAV73567.1 GL450581 EFN80780.1 KZ288215 PBC32452.1 KK852680 KDR18638.1 KK107207 EZA55420.1 GL887707 EGI70313.1 KQ979657 KYN19924.1 NEVH01025149 PNF15681.1 ADTU01007024 GEDC01028010 GEDC01007069 GEDC01005742 GEDC01004929 JAS09288.1 JAS30229.1 JAS31556.1 JAS32369.1 KQ414727 KOC62580.1 JH431954 GECZ01017707 GECZ01000369 JAS52062.1 JAS69400.1 GEBQ01028724 JAT11253.1 GFJQ02007448 JAV99521.1 KK120358 KFM78113.1 KB631740 ERL85700.1 GAKT01000032 JAA93030.1 GBHO01010157 GBHO01009352 GBRD01013430 GBRD01013429 GDHC01013500 JAG33447.1 JAG34252.1 JAG52396.1 JAQ05129.1 GACK01003246 JAA61788.1 KQ434819 KZC06981.1 GADI01007080 JAA66728.1 GFPF01007010 MAA18156.1 GEDV01005330 JAP83227.1 GBBI01002165 JAC16547.1 KC968967 AHF48775.1 APGK01054189 KB741247 ENN72061.1 KK854236 PTY13548.1 GBBM01001915 JAC33503.1 LNIX01000004 OXA55420.1 GBBK01003131 JAC21351.1 GL732594 EFX72953.1 KQ976885 KYN07279.1 LJIJ01001352 ODM92030.1 GDIQ01205719 GDIP01048057 LRGB01000704 JAK46006.1 JAM55658.1 KZS16510.1 GDIQ01254961 JAJ96763.1 CAEY01000828 CH477523 EAT39563.1 JXUM01019738 KQ560553 KXJ81915.1 MNPL01001638 OQR78948.1 ATLV01024045 KE525348 KFB50439.1 KQ425206 KOF69770.1 GFDL01010612 JAV24433.1 ABLF02032229 ABLF02032230 DS231813 EDS25688.1 GGFL01004276 MBW68454.1 GDIQ01120593 JAL31133.1 GDIQ01270580 JAJ81144.1 GFXV01002121 MBW13926.1 AXCM01003669 GGMS01004945 MBY74148.1 GGFK01010763 MBW44084.1 AJVK01011076 AXCN02000516 AAAB01008960 EAA10719.4 APCN01001491 JXLN01015107 KPM10409.1 EF070581 ABM55647.1 AM421517 AM421518 CAM07121.1 AMQN01007448 AMQN01007449 KB300721 ELU06364.1 ADMH02002165 ETN58106.1 AMQM01001448 AMQM01001449 KB097495 ESN96676.1 GFAC01000422 JAT98766.1 BT077759 ACO12183.1 GDAI01002284 JAI15319.1 GANP01001614 JAB82854.1 UYRR01032269 VDK54882.1 GDIP01135909 JAL67805.1 JH818012 EKC38515.1 GDIQ01147947 JAL03779.1 GDIP01223297 JAJ00105.1 JI169341 ADY43821.1 GFWV01009669 MAA34398.1 IACF01002577 LAB68225.1 LBMM01001134 KMQ96849.1
KQ459185 KPJ03298.1 BABH01021571 GQ250576 ACT83401.1 ODYU01000850 SOQ36234.1 AGBW02012080 OWR45711.1 JTDY01000700 KOB76173.1 KQ971354 EFA06801.2 QOIP01000002 RLU26207.1 GEZM01054547 JAV73567.1 GL450581 EFN80780.1 KZ288215 PBC32452.1 KK852680 KDR18638.1 KK107207 EZA55420.1 GL887707 EGI70313.1 KQ979657 KYN19924.1 NEVH01025149 PNF15681.1 ADTU01007024 GEDC01028010 GEDC01007069 GEDC01005742 GEDC01004929 JAS09288.1 JAS30229.1 JAS31556.1 JAS32369.1 KQ414727 KOC62580.1 JH431954 GECZ01017707 GECZ01000369 JAS52062.1 JAS69400.1 GEBQ01028724 JAT11253.1 GFJQ02007448 JAV99521.1 KK120358 KFM78113.1 KB631740 ERL85700.1 GAKT01000032 JAA93030.1 GBHO01010157 GBHO01009352 GBRD01013430 GBRD01013429 GDHC01013500 JAG33447.1 JAG34252.1 JAG52396.1 JAQ05129.1 GACK01003246 JAA61788.1 KQ434819 KZC06981.1 GADI01007080 JAA66728.1 GFPF01007010 MAA18156.1 GEDV01005330 JAP83227.1 GBBI01002165 JAC16547.1 KC968967 AHF48775.1 APGK01054189 KB741247 ENN72061.1 KK854236 PTY13548.1 GBBM01001915 JAC33503.1 LNIX01000004 OXA55420.1 GBBK01003131 JAC21351.1 GL732594 EFX72953.1 KQ976885 KYN07279.1 LJIJ01001352 ODM92030.1 GDIQ01205719 GDIP01048057 LRGB01000704 JAK46006.1 JAM55658.1 KZS16510.1 GDIQ01254961 JAJ96763.1 CAEY01000828 CH477523 EAT39563.1 JXUM01019738 KQ560553 KXJ81915.1 MNPL01001638 OQR78948.1 ATLV01024045 KE525348 KFB50439.1 KQ425206 KOF69770.1 GFDL01010612 JAV24433.1 ABLF02032229 ABLF02032230 DS231813 EDS25688.1 GGFL01004276 MBW68454.1 GDIQ01120593 JAL31133.1 GDIQ01270580 JAJ81144.1 GFXV01002121 MBW13926.1 AXCM01003669 GGMS01004945 MBY74148.1 GGFK01010763 MBW44084.1 AJVK01011076 AXCN02000516 AAAB01008960 EAA10719.4 APCN01001491 JXLN01015107 KPM10409.1 EF070581 ABM55647.1 AM421517 AM421518 CAM07121.1 AMQN01007448 AMQN01007449 KB300721 ELU06364.1 ADMH02002165 ETN58106.1 AMQM01001448 AMQM01001449 KB097495 ESN96676.1 GFAC01000422 JAT98766.1 BT077759 ACO12183.1 GDAI01002284 JAI15319.1 GANP01001614 JAB82854.1 UYRR01032269 VDK54882.1 GDIP01135909 JAL67805.1 JH818012 EKC38515.1 GDIQ01147947 JAL03779.1 GDIP01223297 JAJ00105.1 JI169341 ADY43821.1 GFWV01009669 MAA34398.1 IACF01002577 LAB68225.1 LBMM01001134 KMQ96849.1
Proteomes
UP000218220
UP000053240
UP000283053
UP000053268
UP000005204
UP000007151
+ More
UP000037510 UP000007266 UP000192223 UP000279307 UP000008237 UP000002358 UP000242457 UP000005203 UP000027135 UP000053097 UP000007755 UP000078492 UP000235965 UP000005205 UP000053825 UP000079169 UP000054359 UP000030742 UP000076502 UP000019118 UP000198287 UP000000305 UP000078542 UP000094527 UP000076858 UP000015104 UP000008820 UP000069940 UP000249989 UP000192247 UP000030765 UP000053454 UP000007819 UP000002320 UP000075880 UP000069272 UP000075883 UP000075920 UP000085678 UP000075902 UP000092462 UP000076408 UP000075886 UP000075903 UP000075884 UP000075881 UP000075882 UP000076407 UP000007062 UP000075840 UP000014760 UP000000673 UP000015101 UP000036680 UP000005408 UP000036403
UP000037510 UP000007266 UP000192223 UP000279307 UP000008237 UP000002358 UP000242457 UP000005203 UP000027135 UP000053097 UP000007755 UP000078492 UP000235965 UP000005205 UP000053825 UP000079169 UP000054359 UP000030742 UP000076502 UP000019118 UP000198287 UP000000305 UP000078542 UP000094527 UP000076858 UP000015104 UP000008820 UP000069940 UP000249989 UP000192247 UP000030765 UP000053454 UP000007819 UP000002320 UP000075880 UP000069272 UP000075883 UP000075920 UP000085678 UP000075902 UP000092462 UP000076408 UP000075886 UP000075903 UP000075884 UP000075881 UP000075882 UP000076407 UP000007062 UP000075840 UP000014760 UP000000673 UP000015101 UP000036680 UP000005408 UP000036403
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
ProteinModelPortal
A0A2A4JBP9
A0A194QVV7
A0A3S2PCR7
A0A194QCN1
D2IGZ4
A0A2H1V7L0
+ More
A0A212EW69 A0A0L7LKW2 D6WPE8 A0A1W4WYE1 A0A3L8E0L8 A0A1Y1LRB6 E2BU63 K7J0W3 A0A2A3EMF0 A0A087ZW58 A0A067RHK8 A0A026WHB2 F4W6H8 A0A195E4Z0 A0A2J7PH71 A0A158P285 A0A1B6C720 A0A0L7QVC5 T1J8G8 A0A1B6FPD9 A0A1B6KII7 A0A1S3D7F6 A0A1Z5KWL9 A0A087UL74 U4U6W6 T1DCI9 A0A0A9YMH8 L7MEV1 A0A154P546 A0A0K8R794 A0A224YKK2 A0A131YW97 A0A023F5U0 W0G1U1 N6TUL8 A0A2R7W298 A0A023GKE3 A0A226EDC4 A0A023FHB4 E9H5F3 A0A195D4C1 A0A1D2MGB3 A0A0P5J3W9 A0A0P5FHT3 T1JXF9 Q16Y76 A0A182GVY2 A0A1V9Y000 A0A084WJP5 A0A0L8FZI1 A0A1Q3FAB3 J9JP19 B0VZB2 A0A182J8Q3 A0A2M4CSX8 A0A0P5QQZ8 A0A182FHV6 A0A0P5ET06 A0A2H8TII6 A0A182MB04 A0A182WQB3 A0A2S2Q8T2 A0A2M4ATJ1 A0A1S3JI38 A0A182U3P4 A0A1B0D3W9 A0A182YSP5 A0A182QG78 A0A182V432 A0A182NJR3 A0A182JXS9 A0A182L359 A0A182XML7 Q7Q6L9 A0A182IG84 A0A132AHX3 A2I478 D4FUN3 R7UIH3 W5J2U2 T1FZB4 A0A1E1XI53 C1BT30 A0A0K8TLT7 V5HW58 A0A0M3K4Q6 A0A0P5T8L2 K1RBB0 A0A0N8BSY9 A0A0N7ZQ80 F1L117 A0A293M2Z1 A0A2P2I2G9 A0A0J7L331
A0A212EW69 A0A0L7LKW2 D6WPE8 A0A1W4WYE1 A0A3L8E0L8 A0A1Y1LRB6 E2BU63 K7J0W3 A0A2A3EMF0 A0A087ZW58 A0A067RHK8 A0A026WHB2 F4W6H8 A0A195E4Z0 A0A2J7PH71 A0A158P285 A0A1B6C720 A0A0L7QVC5 T1J8G8 A0A1B6FPD9 A0A1B6KII7 A0A1S3D7F6 A0A1Z5KWL9 A0A087UL74 U4U6W6 T1DCI9 A0A0A9YMH8 L7MEV1 A0A154P546 A0A0K8R794 A0A224YKK2 A0A131YW97 A0A023F5U0 W0G1U1 N6TUL8 A0A2R7W298 A0A023GKE3 A0A226EDC4 A0A023FHB4 E9H5F3 A0A195D4C1 A0A1D2MGB3 A0A0P5J3W9 A0A0P5FHT3 T1JXF9 Q16Y76 A0A182GVY2 A0A1V9Y000 A0A084WJP5 A0A0L8FZI1 A0A1Q3FAB3 J9JP19 B0VZB2 A0A182J8Q3 A0A2M4CSX8 A0A0P5QQZ8 A0A182FHV6 A0A0P5ET06 A0A2H8TII6 A0A182MB04 A0A182WQB3 A0A2S2Q8T2 A0A2M4ATJ1 A0A1S3JI38 A0A182U3P4 A0A1B0D3W9 A0A182YSP5 A0A182QG78 A0A182V432 A0A182NJR3 A0A182JXS9 A0A182L359 A0A182XML7 Q7Q6L9 A0A182IG84 A0A132AHX3 A2I478 D4FUN3 R7UIH3 W5J2U2 T1FZB4 A0A1E1XI53 C1BT30 A0A0K8TLT7 V5HW58 A0A0M3K4Q6 A0A0P5T8L2 K1RBB0 A0A0N8BSY9 A0A0N7ZQ80 F1L117 A0A293M2Z1 A0A2P2I2G9 A0A0J7L331
PDB
4AU8
E-value=1.21531e-142,
Score=1297
Ontologies
GO
Topology
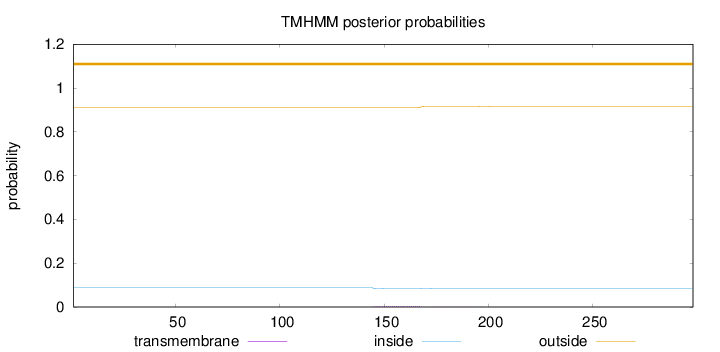
Length:
298
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10206
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08855
outside
1 - 298
Population Genetic Test Statistics
Pi
228.621345
Theta
213.133693
Tajima's D
-0.819312
CLR
2.059942
CSRT
0.171541422928854
Interpretation
Uncertain
