Gene
KWMTBOMO11591
Pre Gene Modal
BGIBMGA004100
Annotation
PREDICTED:_DNA_replication_complex_GINS_protein_PSF3-like_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 2.055
Sequence
CDS
ATGGGATTCTTAGACCCTTCTGCTGCTGATGTTGACCTAAAAGCCGGCTGCAGTGTAGAAATACCACTATGGCTAGCCGAATCCTTATATTCTAGAAGACCTCCTCTGGTTAGCGTAGAATTGCCAAAAATCTATAAAGAATCATATAGAGAGATCCTAAATGCGGATGCGTGCGCGGTCGATCTTCATAAGTTAGGGCAGCATTTCTATGAACTTGGATGCTATGTAGCTAAACATGATATCAAAAGCGAAGTTGCTGCTACATTAAACAATACATATCGTCAAAGGTTTAGAATGTTATTGGCTGCCAGTATGTCGAGTGACTCAATAAACACAATGCAACCACTTTCTGCTAGTGAGAGAATACAAGCTGCAGATGCTTCTAATACTGAGAGATCATTTTTAACCTGGTTACAAAGAGGAGACACTCCATTGACAACAGCTAATATGGTTGCAAATCACAGGAAAAGAAAGCGTGCTGAAATGGAAATGCTATAG
Protein
MGFLDPSAADVDLKAGCSVEIPLWLAESLYSRRPPLVSVELPKIYKESYREILNADACAVDLHKLGQHFYELGCYVAKHDIKSEVAATLNNTYRQRFRMLLAASMSSDSINTMQPLSASERIQAADASNTERSFLTWLQRGDTPLTTANMVANHRKRKRAEMEML
Summary
Uniprot
H9J3L3
A0A2A4JDK7
S4PPM5
A0A194QVP6
A0A194QD03
A0A212EW42
+ More
A0A2H1V780 A0A0L7LLV9 A0A2J7PI27 A0A1Y1JX65 A0A0T6B384 A0A067R1C6 A0A2P6L1W0 D6WSJ1 N6TBT3 A0A2P8Z3A5 A0A1W4XJU6 A7SMZ3 A0A2B4RIA1 A0A3M6U830 A0A210QQ66 K1RMP1 A0A087T995 W4YRM4 A0A1S3GZP2 E9H7Y6 A0A154NXX7 A0A1S3KBC5 C3XTX5 A0A0P5TE53 A0A0P5TJA1 A0A232FGI0 A0A0P5XMB3 A0A1J1ID14 V3ZGB8 K7IST4 T1JJC2 E2C294 A0A0P5FHR7 A0A3L5TQ06 A0A1A8L8V5 A0A1A8UWN4 F4WYZ6 A0A3S0Z9C2 A0A2R5LH68 E9ILY8 E1ZVI3 A0A195F9T0 A0A195CEN8 A0A1B6G233 A0A0B7BGQ5 A0A3N0Z7Y2 A0A1B6I7M0 A0A026WVT8 A0A195BWZ9 A0A3Q2X4D5 A0A1B6DKP1 A0A3B4H0C4 A0A3P8NH50 A0A3P9CGB3 A0A158NC88 A0A147BKE5 A0A2A3E180 A0A3Q3M5G1 T1FNF9 A0A3P9IM13 A0A131Y8R3 A0A3Q2DDS4 A0A0N8AC23 A0A3B3INW2 A0A1B6C2G2 A0A1B6M091 A0A3Q0RNQ6 A0A3B4TB60 W5M6E5 A0A3S2PE49 E3TET6 A0A0N8JVY7 A0A1W4YTE8 A0A3B4WX09 A0A3Q2ZW21 A0A3Q2PZ89 I3JXP0 A0A2P2I283 A0A3M7SVQ4 A0A2I4D9J0 A0A195EBD9 A0A3B3DDL4 A0A151WFP2 A0A3Q2ZFU0 Q7ZT01 A0A2U9C105 G3NCE0 A0A060X294 A0A1S3KMJ2 A0A3B3DBK4 A0A3B1K740 A0A3B1JTL2 A0A3Q3QG96 A0A1U7QGW5
A0A2H1V780 A0A0L7LLV9 A0A2J7PI27 A0A1Y1JX65 A0A0T6B384 A0A067R1C6 A0A2P6L1W0 D6WSJ1 N6TBT3 A0A2P8Z3A5 A0A1W4XJU6 A7SMZ3 A0A2B4RIA1 A0A3M6U830 A0A210QQ66 K1RMP1 A0A087T995 W4YRM4 A0A1S3GZP2 E9H7Y6 A0A154NXX7 A0A1S3KBC5 C3XTX5 A0A0P5TE53 A0A0P5TJA1 A0A232FGI0 A0A0P5XMB3 A0A1J1ID14 V3ZGB8 K7IST4 T1JJC2 E2C294 A0A0P5FHR7 A0A3L5TQ06 A0A1A8L8V5 A0A1A8UWN4 F4WYZ6 A0A3S0Z9C2 A0A2R5LH68 E9ILY8 E1ZVI3 A0A195F9T0 A0A195CEN8 A0A1B6G233 A0A0B7BGQ5 A0A3N0Z7Y2 A0A1B6I7M0 A0A026WVT8 A0A195BWZ9 A0A3Q2X4D5 A0A1B6DKP1 A0A3B4H0C4 A0A3P8NH50 A0A3P9CGB3 A0A158NC88 A0A147BKE5 A0A2A3E180 A0A3Q3M5G1 T1FNF9 A0A3P9IM13 A0A131Y8R3 A0A3Q2DDS4 A0A0N8AC23 A0A3B3INW2 A0A1B6C2G2 A0A1B6M091 A0A3Q0RNQ6 A0A3B4TB60 W5M6E5 A0A3S2PE49 E3TET6 A0A0N8JVY7 A0A1W4YTE8 A0A3B4WX09 A0A3Q2ZW21 A0A3Q2PZ89 I3JXP0 A0A2P2I283 A0A3M7SVQ4 A0A2I4D9J0 A0A195EBD9 A0A3B3DDL4 A0A151WFP2 A0A3Q2ZFU0 Q7ZT01 A0A2U9C105 G3NCE0 A0A060X294 A0A1S3KMJ2 A0A3B3DBK4 A0A3B1K740 A0A3B1JTL2 A0A3Q3QG96 A0A1U7QGW5
Pubmed
19121390
23622113
26354079
22118469
26227816
28004739
+ More
24845553 18362917 19820115 23537049 29403074 17615350 30382153 28812685 22992520 21292972 18563158 28648823 23254933 20075255 20798317 26977809 21719571 21282665 24508170 30249741 25186727 21347285 29652888 17554307 20634964 30375419 29451363 12730133 24755649 25329095
24845553 18362917 19820115 23537049 29403074 17615350 30382153 28812685 22992520 21292972 18563158 28648823 23254933 20075255 20798317 26977809 21719571 21282665 24508170 30249741 25186727 21347285 29652888 17554307 20634964 30375419 29451363 12730133 24755649 25329095
EMBL
BABH01021571
NWSH01001992
PCG69493.1
GAIX01000157
JAA92403.1
KQ461073
+ More
KPJ09608.1 KQ459185 KPJ03299.1 AGBW02012080 OWR45710.1 ODYU01000850 SOQ36232.1 JTDY01000700 KOB76171.1 NEVH01025130 PNF15981.1 GEZM01100694 JAV52818.1 LJIG01016026 KRT81836.1 KK852779 KDR16687.1 MWRG01002486 PRD32556.1 KQ971354 EFA07113.1 APGK01036196 KB740937 KB632052 ENN77754.1 ERL88332.1 PYGN01000217 PSN50963.1 DS469715 EDO34905.1 LSMT01000538 PFX16529.1 RCHS01002051 RMX49823.1 NEDP02002420 OWF50859.1 JH817099 EKC42875.1 KK114081 KFM61684.1 AAGJ04146738 GL732602 EFX72111.1 KQ434781 KZC04467.1 GG666464 EEN68349.1 GDIP01127764 JAL75950.1 GDIP01131228 LRGB01000024 JAL72486.1 KZS21766.1 NNAY01000238 OXU29795.1 GDIP01070018 JAM33697.1 CVRI01000047 CRK98175.1 KB202444 ESO90268.1 AAZX01006436 JH431448 GL452091 EFN77941.1 GDIQ01257824 JAJ93900.1 KV593078 OPL21288.1 HAEF01003541 SBR40923.1 HADY01009705 HAEJ01011285 SBS51742.1 GL888463 EGI60650.1 RQTK01001581 RUS69771.1 GGLE01004744 MBY08870.1 GL764129 EFZ18298.1 GL434534 EFN74820.1 KQ981727 KYN37136.1 KQ977873 KYM99165.1 GECZ01013416 JAS56353.1 HACG01045182 CEK92047.1 RJVU01006614 ROL54560.1 GECU01024803 JAS82903.1 KK107079 QOIP01000010 EZA60150.1 RLU17374.1 KQ976396 KYM92815.1 GEDC01011047 JAS26251.1 ADTU01011743 GEGO01004154 JAR91250.1 KZ288476 PBC25284.1 AMQM01006392 KB097417 ESN97202.1 GEFM01001310 JAP74486.1 GDIP01162153 JAJ61249.1 GEDC01029585 GEDC01018926 JAS07713.1 JAS18372.1 GEBQ01010636 JAT29341.1 AHAT01010569 CM012442 RVE71378.1 GU588868 ADO28822.1 JARO02012126 KPP59449.1 AERX01003511 AERX01003512 IACF01002504 LAB68155.1 REGN01000696 RNA39904.1 KQ979236 KYN22154.1 KQ983211 KYQ46605.1 BC043845 BC106314 AB097170 AAH43845.1 AAI06315.1 BAC66460.1 CP026252 AWP08982.1 FR904913 CDQ73581.1
KPJ09608.1 KQ459185 KPJ03299.1 AGBW02012080 OWR45710.1 ODYU01000850 SOQ36232.1 JTDY01000700 KOB76171.1 NEVH01025130 PNF15981.1 GEZM01100694 JAV52818.1 LJIG01016026 KRT81836.1 KK852779 KDR16687.1 MWRG01002486 PRD32556.1 KQ971354 EFA07113.1 APGK01036196 KB740937 KB632052 ENN77754.1 ERL88332.1 PYGN01000217 PSN50963.1 DS469715 EDO34905.1 LSMT01000538 PFX16529.1 RCHS01002051 RMX49823.1 NEDP02002420 OWF50859.1 JH817099 EKC42875.1 KK114081 KFM61684.1 AAGJ04146738 GL732602 EFX72111.1 KQ434781 KZC04467.1 GG666464 EEN68349.1 GDIP01127764 JAL75950.1 GDIP01131228 LRGB01000024 JAL72486.1 KZS21766.1 NNAY01000238 OXU29795.1 GDIP01070018 JAM33697.1 CVRI01000047 CRK98175.1 KB202444 ESO90268.1 AAZX01006436 JH431448 GL452091 EFN77941.1 GDIQ01257824 JAJ93900.1 KV593078 OPL21288.1 HAEF01003541 SBR40923.1 HADY01009705 HAEJ01011285 SBS51742.1 GL888463 EGI60650.1 RQTK01001581 RUS69771.1 GGLE01004744 MBY08870.1 GL764129 EFZ18298.1 GL434534 EFN74820.1 KQ981727 KYN37136.1 KQ977873 KYM99165.1 GECZ01013416 JAS56353.1 HACG01045182 CEK92047.1 RJVU01006614 ROL54560.1 GECU01024803 JAS82903.1 KK107079 QOIP01000010 EZA60150.1 RLU17374.1 KQ976396 KYM92815.1 GEDC01011047 JAS26251.1 ADTU01011743 GEGO01004154 JAR91250.1 KZ288476 PBC25284.1 AMQM01006392 KB097417 ESN97202.1 GEFM01001310 JAP74486.1 GDIP01162153 JAJ61249.1 GEDC01029585 GEDC01018926 JAS07713.1 JAS18372.1 GEBQ01010636 JAT29341.1 AHAT01010569 CM012442 RVE71378.1 GU588868 ADO28822.1 JARO02012126 KPP59449.1 AERX01003511 AERX01003512 IACF01002504 LAB68155.1 REGN01000696 RNA39904.1 KQ979236 KYN22154.1 KQ983211 KYQ46605.1 BC043845 BC106314 AB097170 AAH43845.1 AAI06315.1 BAC66460.1 CP026252 AWP08982.1 FR904913 CDQ73581.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000235965 UP000027135 UP000007266 UP000019118 UP000030742 UP000245037 UP000192223 UP000001593 UP000225706 UP000275408 UP000242188 UP000005408 UP000054359 UP000007110 UP000085678 UP000000305 UP000076502 UP000001554 UP000076858 UP000215335 UP000183832 UP000030746 UP000002358 UP000008237 UP000007755 UP000271974 UP000000311 UP000078541 UP000078542 UP000053097 UP000279307 UP000078540 UP000264840 UP000261460 UP000265100 UP000265160 UP000005205 UP000242457 UP000261640 UP000015101 UP000265200 UP000265020 UP000001038 UP000261340 UP000261420 UP000018468 UP000221080 UP000034805 UP000192224 UP000261360 UP000264800 UP000265000 UP000005207 UP000276133 UP000192220 UP000078492 UP000261560 UP000075809 UP000264820 UP000246464 UP000007635 UP000193380 UP000087266 UP000018467 UP000261600 UP000189706
UP000235965 UP000027135 UP000007266 UP000019118 UP000030742 UP000245037 UP000192223 UP000001593 UP000225706 UP000275408 UP000242188 UP000005408 UP000054359 UP000007110 UP000085678 UP000000305 UP000076502 UP000001554 UP000076858 UP000215335 UP000183832 UP000030746 UP000002358 UP000008237 UP000007755 UP000271974 UP000000311 UP000078541 UP000078542 UP000053097 UP000279307 UP000078540 UP000264840 UP000261460 UP000265100 UP000265160 UP000005205 UP000242457 UP000261640 UP000015101 UP000265200 UP000265020 UP000001038 UP000261340 UP000261420 UP000018468 UP000221080 UP000034805 UP000192224 UP000261360 UP000264800 UP000265000 UP000005207 UP000276133 UP000192220 UP000078492 UP000261560 UP000075809 UP000264820 UP000246464 UP000007635 UP000193380 UP000087266 UP000018467 UP000261600 UP000189706
PRIDE
Pfam
PF05916 Sld5
Interpro
SUPFAM
SSF158573
SSF158573
Gene 3D
ProteinModelPortal
H9J3L3
A0A2A4JDK7
S4PPM5
A0A194QVP6
A0A194QD03
A0A212EW42
+ More
A0A2H1V780 A0A0L7LLV9 A0A2J7PI27 A0A1Y1JX65 A0A0T6B384 A0A067R1C6 A0A2P6L1W0 D6WSJ1 N6TBT3 A0A2P8Z3A5 A0A1W4XJU6 A7SMZ3 A0A2B4RIA1 A0A3M6U830 A0A210QQ66 K1RMP1 A0A087T995 W4YRM4 A0A1S3GZP2 E9H7Y6 A0A154NXX7 A0A1S3KBC5 C3XTX5 A0A0P5TE53 A0A0P5TJA1 A0A232FGI0 A0A0P5XMB3 A0A1J1ID14 V3ZGB8 K7IST4 T1JJC2 E2C294 A0A0P5FHR7 A0A3L5TQ06 A0A1A8L8V5 A0A1A8UWN4 F4WYZ6 A0A3S0Z9C2 A0A2R5LH68 E9ILY8 E1ZVI3 A0A195F9T0 A0A195CEN8 A0A1B6G233 A0A0B7BGQ5 A0A3N0Z7Y2 A0A1B6I7M0 A0A026WVT8 A0A195BWZ9 A0A3Q2X4D5 A0A1B6DKP1 A0A3B4H0C4 A0A3P8NH50 A0A3P9CGB3 A0A158NC88 A0A147BKE5 A0A2A3E180 A0A3Q3M5G1 T1FNF9 A0A3P9IM13 A0A131Y8R3 A0A3Q2DDS4 A0A0N8AC23 A0A3B3INW2 A0A1B6C2G2 A0A1B6M091 A0A3Q0RNQ6 A0A3B4TB60 W5M6E5 A0A3S2PE49 E3TET6 A0A0N8JVY7 A0A1W4YTE8 A0A3B4WX09 A0A3Q2ZW21 A0A3Q2PZ89 I3JXP0 A0A2P2I283 A0A3M7SVQ4 A0A2I4D9J0 A0A195EBD9 A0A3B3DDL4 A0A151WFP2 A0A3Q2ZFU0 Q7ZT01 A0A2U9C105 G3NCE0 A0A060X294 A0A1S3KMJ2 A0A3B3DBK4 A0A3B1K740 A0A3B1JTL2 A0A3Q3QG96 A0A1U7QGW5
A0A2H1V780 A0A0L7LLV9 A0A2J7PI27 A0A1Y1JX65 A0A0T6B384 A0A067R1C6 A0A2P6L1W0 D6WSJ1 N6TBT3 A0A2P8Z3A5 A0A1W4XJU6 A7SMZ3 A0A2B4RIA1 A0A3M6U830 A0A210QQ66 K1RMP1 A0A087T995 W4YRM4 A0A1S3GZP2 E9H7Y6 A0A154NXX7 A0A1S3KBC5 C3XTX5 A0A0P5TE53 A0A0P5TJA1 A0A232FGI0 A0A0P5XMB3 A0A1J1ID14 V3ZGB8 K7IST4 T1JJC2 E2C294 A0A0P5FHR7 A0A3L5TQ06 A0A1A8L8V5 A0A1A8UWN4 F4WYZ6 A0A3S0Z9C2 A0A2R5LH68 E9ILY8 E1ZVI3 A0A195F9T0 A0A195CEN8 A0A1B6G233 A0A0B7BGQ5 A0A3N0Z7Y2 A0A1B6I7M0 A0A026WVT8 A0A195BWZ9 A0A3Q2X4D5 A0A1B6DKP1 A0A3B4H0C4 A0A3P8NH50 A0A3P9CGB3 A0A158NC88 A0A147BKE5 A0A2A3E180 A0A3Q3M5G1 T1FNF9 A0A3P9IM13 A0A131Y8R3 A0A3Q2DDS4 A0A0N8AC23 A0A3B3INW2 A0A1B6C2G2 A0A1B6M091 A0A3Q0RNQ6 A0A3B4TB60 W5M6E5 A0A3S2PE49 E3TET6 A0A0N8JVY7 A0A1W4YTE8 A0A3B4WX09 A0A3Q2ZW21 A0A3Q2PZ89 I3JXP0 A0A2P2I283 A0A3M7SVQ4 A0A2I4D9J0 A0A195EBD9 A0A3B3DDL4 A0A151WFP2 A0A3Q2ZFU0 Q7ZT01 A0A2U9C105 G3NCE0 A0A060X294 A0A1S3KMJ2 A0A3B3DBK4 A0A3B1K740 A0A3B1JTL2 A0A3Q3QG96 A0A1U7QGW5
PDB
2Q9Q
E-value=5.06898e-21,
Score=244
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
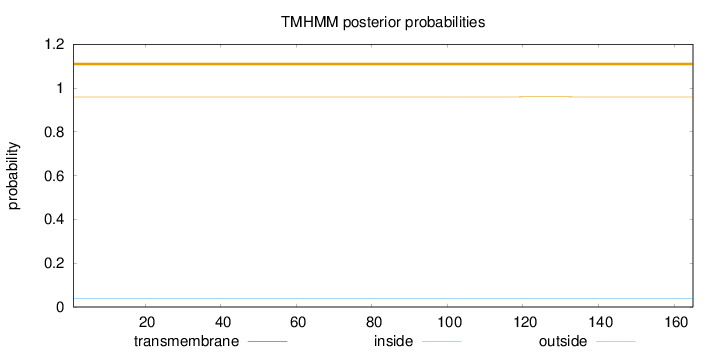
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00116
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.04035
outside
1 - 165
Population Genetic Test Statistics
Pi
109.155581
Theta
148.121706
Tajima's D
-0.843473
CLR
27.859257
CSRT
0.170941452927354
Interpretation
Uncertain
