Pre Gene Modal
BGIBMGA004099
Annotation
PREDICTED:_TM2_domain-containing_protein_almondex_[Amyelois_transitella]
Full name
TM2 domain-containing protein almondex
Location in the cell
Extracellular Reliability : 2.201
Sequence
CDS
ATGGCTAATAATAACATGGATTTTCATAGTAAAGAGAATAAAATAAAAGGTGATTCAAATGACAAAGGTGTAAGTTTTAGTGAGAATGAAGAATACATAAAAAGTTGTCCATCAGTTGATCAAGAGAATATGACTGAATGTTCCTCTTTACTGTACCCTTGTATAACATGTAACCGGAGTATAGATTGCAGATATGGAGGTTTCTATAATTATACATGTACAGTGAAACCAAAAGTAGTTTGTCAAGGGTTAAAAACTTTCAACAAAAGCTCAATGTGTAGATTTTGCTATCAAACTGATAAATGGGAACATCGTTGTGAACAAAAGGCAAATTGTAATTCTTTAGCTTCACCTCTCAAATACTATCTAACTAATTGCACTGTATCTGATGATGTTATATGTTTGGGGAAGAGGCGGTTCCTTAAGAAAGTAAGATGCTCCTGGACATCAGGAACTCGTTGGGGTACAGCACTTGTTCTGGCATTAACTTTAGGAGGTTTTGGAGCTGACCGCTTTTATCTTGGTCATTGGCAAGAAGGCATTGGTAAACTTTTTAGTTTTGGTGGCCTTGGAGTGTGGACTCTAGTGGATGCATTGTTAATTGGAATACACCATTTAGGACCAGCTGATGGGTCTTTATATATTTAA
Protein
MANNNMDFHSKENKIKGDSNDKGVSFSENEEYIKSCPSVDQENMTECSSLLYPCITCNRSIDCRYGGFYNYTCTVKPKVVCQGLKTFNKSSMCRFCYQTDKWEHRCEQKANCNSLASPLKYYLTNCTVSDDVICLGKRRFLKKVRCSWTSGTRWGTALVLALTLGGFGADRFYLGHWQEGIGKLFSFGGLGVWTLVDALLIGIHHLGPADGSLYI
Summary
Similarity
Belongs to the TM2 family.
Keywords
Complete proteome
Developmental protein
Glycoprotein
Membrane
Notch signaling pathway
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain TM2 domain-containing protein almondex
Uniprot
A0A2A4JGJ8
A0A2H1VUB0
I4DJI1
A0A194QXC7
A0A194QIM1
A0A0N1IN80
+ More
A0A3S2LZL4 A0A0L7LLF6 A0A212FP06 S4PSS1 A0A1Y1M004 A0A195F485 E2BSI7 E9J329 A0A195D9X0 A0A151IEY8 A0A026X013 A0A151XEI1 A0A0T6AWF3 A0A0J7NT61 A0A154PLX8 B4JKN1 A0A088A2P6 A0A2A3EH38 A0A0L7RB87 A0A0C9QXG6 A0A1S3D0W0 A0A0K8TP88 B4M1R6 A0A1W4X925 A0A139WER1 A0A0N0U5E8 C4WSW3 B4L8S4 A0A0M3QZC1 A0A1J1IBA0 B4NC55 A0A0K8VTR4 K7J5W9 A0A034V6N8 B3MZN3 A0A2S2QKY2 A0A067RF48 A0A1B0GLA7 A0A084WRL8 A0A1I8P3S3 A0A069DQH3 A0A3B0K7E0 B4H4N0 A0A0L0C4G1 A0A1B6CCP3 A0A2H8TPK8 J3JUM2 U5EWB0 A0A023EL58 A0A1W7R511 A0A1W4VHH0 A0A1L8D9H1 A0A224XYX6 E0VEV3 A0A1B6IG51 A0A1B6GF21 A0A0A9W7P8 A0A1I8MY21 A0A2M4AW38 Q16J55 M9NF00 Q9U4H5 B4PWU5 B4ILP3 B3NT89 A0A182XP59 A0A2S2P3F1 Q7Q602 A0A182TV90 B4NU72 T1J3U4 A0A182JZ66 A0A182WHV1 A0A182RGT1 W5J4D3 A0A182QWN4 A0A182NLQ7 A0A1S4GRB1 A0A182KP22 A0A182P108 A0A182LZQ8 A0A1B0FQY6 A0A182I1G5 D3TQ17 A0A1A9XXP5 A0A182GVL7 A0A182V9W8 T1GXD3 A0A182JC26 A0A182FEU9 A0A1B0ADQ5 Q29HX1 A0A1A9UEL1
A0A3S2LZL4 A0A0L7LLF6 A0A212FP06 S4PSS1 A0A1Y1M004 A0A195F485 E2BSI7 E9J329 A0A195D9X0 A0A151IEY8 A0A026X013 A0A151XEI1 A0A0T6AWF3 A0A0J7NT61 A0A154PLX8 B4JKN1 A0A088A2P6 A0A2A3EH38 A0A0L7RB87 A0A0C9QXG6 A0A1S3D0W0 A0A0K8TP88 B4M1R6 A0A1W4X925 A0A139WER1 A0A0N0U5E8 C4WSW3 B4L8S4 A0A0M3QZC1 A0A1J1IBA0 B4NC55 A0A0K8VTR4 K7J5W9 A0A034V6N8 B3MZN3 A0A2S2QKY2 A0A067RF48 A0A1B0GLA7 A0A084WRL8 A0A1I8P3S3 A0A069DQH3 A0A3B0K7E0 B4H4N0 A0A0L0C4G1 A0A1B6CCP3 A0A2H8TPK8 J3JUM2 U5EWB0 A0A023EL58 A0A1W7R511 A0A1W4VHH0 A0A1L8D9H1 A0A224XYX6 E0VEV3 A0A1B6IG51 A0A1B6GF21 A0A0A9W7P8 A0A1I8MY21 A0A2M4AW38 Q16J55 M9NF00 Q9U4H5 B4PWU5 B4ILP3 B3NT89 A0A182XP59 A0A2S2P3F1 Q7Q602 A0A182TV90 B4NU72 T1J3U4 A0A182JZ66 A0A182WHV1 A0A182RGT1 W5J4D3 A0A182QWN4 A0A182NLQ7 A0A1S4GRB1 A0A182KP22 A0A182P108 A0A182LZQ8 A0A1B0FQY6 A0A182I1G5 D3TQ17 A0A1A9XXP5 A0A182GVL7 A0A182V9W8 T1GXD3 A0A182JC26 A0A182FEU9 A0A1B0ADQ5 Q29HX1 A0A1A9UEL1
Pubmed
22651552
26354079
26227816
22118469
23622113
28004739
+ More
20798317 21282665 24508170 30249741 17994087 26369729 18362917 19820115 20075255 25348373 24845553 24438588 26334808 26108605 22516182 23537049 24945155 20566863 25401762 26823975 25315136 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 14595834 10731138 17550304 12364791 20920257 23761445 20966253 20353571 26483478 15632085
20798317 21282665 24508170 30249741 17994087 26369729 18362917 19820115 20075255 25348373 24845553 24438588 26334808 26108605 22516182 23537049 24945155 20566863 25401762 26823975 25315136 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 14595834 10731138 17550304 12364791 20920257 23761445 20966253 20353571 26483478 15632085
EMBL
NWSH01001566
PCG70838.1
ODYU01004206
SOQ43824.1
AK401449
BAM18071.1
+ More
KQ461073 KPJ09610.1 KQ459185 KPJ03301.1 KQ458556 KPJ05821.1 RSAL01000096 RVE47709.1 JTDY01000700 KOB76174.1 AGBW02003976 OWR55484.1 GAIX01013298 JAA79262.1 GEZM01044330 JAV78308.1 KQ981820 KYN35196.1 GL450240 EFN81320.1 GL768055 EFZ12810.1 KQ981082 KYN09657.1 KQ977830 KYM99409.1 KK107054 QOIP01000004 EZA61597.1 RLU24023.1 KQ982254 KYQ58730.1 LJIG01022653 KRT79430.1 LBMM01001895 KMQ95590.1 KQ434977 KZC12859.1 CH916370 EDW00134.1 KZ288252 PBC31028.1 KQ414617 KOC68134.1 GBYB01008479 JAG78246.1 GDAI01001642 JAI15961.1 CH940651 EDW65620.2 KQ971354 KYB26426.1 KQ435793 KOX74228.1 ABLF02023794 AK340358 BAH70983.1 CH933815 EDW08049.1 CP012528 ALC49163.1 CVRI01000043 CRK96254.1 CH964239 EDW82414.1 GDHF01010071 JAI42243.1 GAKP01019961 JAC38991.1 CH902635 EDV33834.1 GGMS01009130 MBY78333.1 KK852727 KDR17617.1 AJWK01034704 ATLV01026170 KE525407 KFB52862.1 GBGD01002973 JAC85916.1 OUUW01000015 SPP88592.1 CH479209 EDW32642.1 JRES01000996 KNC26334.1 GEDC01026086 JAS11212.1 GFXV01003373 MBW15178.1 APGK01034947 BT126937 KB740923 KB632430 AEE61899.1 ENN78099.1 ERL95493.1 GANO01001500 JAB58371.1 GAPW01003650 JAC09948.1 GEHC01001396 JAV46249.1 GFDF01011104 JAV02980.1 GFTR01003185 JAW13241.1 DS235099 EEB11909.1 GECU01021807 JAS85899.1 GECZ01008735 JAS61034.1 GBHO01039785 GBRD01012893 GDHC01006481 JAG03819.1 JAG52933.1 JAQ12148.1 GGFK01011660 MBW44981.1 CH478031 EAT34293.1 AE014298 AFH07305.1 AF217797 AF181623 CM000162 EDX02829.1 CH480883 EDW54750.1 CH954180 EDV46269.1 GGMR01011392 MBY24011.1 AAAB01008960 EAA10917.4 CH983714 EDX16519.1 JH431831 ADMH02002146 ETN58268.1 AXCN02000969 AXCM01001410 CCAG010018611 APCN01000058 EZ423519 ADD19795.1 JXUM01091244 KQ563899 KXJ73090.1 CAQQ02374560 CH379064 EAL31636.3
KQ461073 KPJ09610.1 KQ459185 KPJ03301.1 KQ458556 KPJ05821.1 RSAL01000096 RVE47709.1 JTDY01000700 KOB76174.1 AGBW02003976 OWR55484.1 GAIX01013298 JAA79262.1 GEZM01044330 JAV78308.1 KQ981820 KYN35196.1 GL450240 EFN81320.1 GL768055 EFZ12810.1 KQ981082 KYN09657.1 KQ977830 KYM99409.1 KK107054 QOIP01000004 EZA61597.1 RLU24023.1 KQ982254 KYQ58730.1 LJIG01022653 KRT79430.1 LBMM01001895 KMQ95590.1 KQ434977 KZC12859.1 CH916370 EDW00134.1 KZ288252 PBC31028.1 KQ414617 KOC68134.1 GBYB01008479 JAG78246.1 GDAI01001642 JAI15961.1 CH940651 EDW65620.2 KQ971354 KYB26426.1 KQ435793 KOX74228.1 ABLF02023794 AK340358 BAH70983.1 CH933815 EDW08049.1 CP012528 ALC49163.1 CVRI01000043 CRK96254.1 CH964239 EDW82414.1 GDHF01010071 JAI42243.1 GAKP01019961 JAC38991.1 CH902635 EDV33834.1 GGMS01009130 MBY78333.1 KK852727 KDR17617.1 AJWK01034704 ATLV01026170 KE525407 KFB52862.1 GBGD01002973 JAC85916.1 OUUW01000015 SPP88592.1 CH479209 EDW32642.1 JRES01000996 KNC26334.1 GEDC01026086 JAS11212.1 GFXV01003373 MBW15178.1 APGK01034947 BT126937 KB740923 KB632430 AEE61899.1 ENN78099.1 ERL95493.1 GANO01001500 JAB58371.1 GAPW01003650 JAC09948.1 GEHC01001396 JAV46249.1 GFDF01011104 JAV02980.1 GFTR01003185 JAW13241.1 DS235099 EEB11909.1 GECU01021807 JAS85899.1 GECZ01008735 JAS61034.1 GBHO01039785 GBRD01012893 GDHC01006481 JAG03819.1 JAG52933.1 JAQ12148.1 GGFK01011660 MBW44981.1 CH478031 EAT34293.1 AE014298 AFH07305.1 AF217797 AF181623 CM000162 EDX02829.1 CH480883 EDW54750.1 CH954180 EDV46269.1 GGMR01011392 MBY24011.1 AAAB01008960 EAA10917.4 CH983714 EDX16519.1 JH431831 ADMH02002146 ETN58268.1 AXCN02000969 AXCM01001410 CCAG010018611 APCN01000058 EZ423519 ADD19795.1 JXUM01091244 KQ563899 KXJ73090.1 CAQQ02374560 CH379064 EAL31636.3
Proteomes
UP000218220
UP000053240
UP000053268
UP000283053
UP000037510
UP000007151
+ More
UP000078541 UP000008237 UP000078492 UP000078542 UP000053097 UP000279307 UP000075809 UP000036403 UP000076502 UP000001070 UP000005203 UP000242457 UP000053825 UP000079169 UP000008792 UP000192223 UP000007266 UP000053105 UP000007819 UP000009192 UP000092553 UP000183832 UP000007798 UP000002358 UP000007801 UP000027135 UP000092461 UP000030765 UP000095300 UP000268350 UP000008744 UP000037069 UP000019118 UP000030742 UP000192221 UP000009046 UP000095301 UP000008820 UP000000803 UP000002282 UP000001292 UP000008711 UP000076407 UP000007062 UP000075902 UP000000304 UP000075881 UP000075920 UP000075900 UP000000673 UP000075886 UP000075884 UP000075882 UP000075885 UP000075883 UP000092444 UP000075840 UP000092443 UP000069940 UP000249989 UP000075903 UP000015102 UP000075880 UP000069272 UP000092445 UP000001819 UP000078200
UP000078541 UP000008237 UP000078492 UP000078542 UP000053097 UP000279307 UP000075809 UP000036403 UP000076502 UP000001070 UP000005203 UP000242457 UP000053825 UP000079169 UP000008792 UP000192223 UP000007266 UP000053105 UP000007819 UP000009192 UP000092553 UP000183832 UP000007798 UP000002358 UP000007801 UP000027135 UP000092461 UP000030765 UP000095300 UP000268350 UP000008744 UP000037069 UP000019118 UP000030742 UP000192221 UP000009046 UP000095301 UP000008820 UP000000803 UP000002282 UP000001292 UP000008711 UP000076407 UP000007062 UP000075902 UP000000304 UP000075881 UP000075920 UP000075900 UP000000673 UP000075886 UP000075884 UP000075882 UP000075885 UP000075883 UP000092444 UP000075840 UP000092443 UP000069940 UP000249989 UP000075903 UP000015102 UP000075880 UP000069272 UP000092445 UP000001819 UP000078200
PRIDE
Interpro
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
A0A2A4JGJ8
A0A2H1VUB0
I4DJI1
A0A194QXC7
A0A194QIM1
A0A0N1IN80
+ More
A0A3S2LZL4 A0A0L7LLF6 A0A212FP06 S4PSS1 A0A1Y1M004 A0A195F485 E2BSI7 E9J329 A0A195D9X0 A0A151IEY8 A0A026X013 A0A151XEI1 A0A0T6AWF3 A0A0J7NT61 A0A154PLX8 B4JKN1 A0A088A2P6 A0A2A3EH38 A0A0L7RB87 A0A0C9QXG6 A0A1S3D0W0 A0A0K8TP88 B4M1R6 A0A1W4X925 A0A139WER1 A0A0N0U5E8 C4WSW3 B4L8S4 A0A0M3QZC1 A0A1J1IBA0 B4NC55 A0A0K8VTR4 K7J5W9 A0A034V6N8 B3MZN3 A0A2S2QKY2 A0A067RF48 A0A1B0GLA7 A0A084WRL8 A0A1I8P3S3 A0A069DQH3 A0A3B0K7E0 B4H4N0 A0A0L0C4G1 A0A1B6CCP3 A0A2H8TPK8 J3JUM2 U5EWB0 A0A023EL58 A0A1W7R511 A0A1W4VHH0 A0A1L8D9H1 A0A224XYX6 E0VEV3 A0A1B6IG51 A0A1B6GF21 A0A0A9W7P8 A0A1I8MY21 A0A2M4AW38 Q16J55 M9NF00 Q9U4H5 B4PWU5 B4ILP3 B3NT89 A0A182XP59 A0A2S2P3F1 Q7Q602 A0A182TV90 B4NU72 T1J3U4 A0A182JZ66 A0A182WHV1 A0A182RGT1 W5J4D3 A0A182QWN4 A0A182NLQ7 A0A1S4GRB1 A0A182KP22 A0A182P108 A0A182LZQ8 A0A1B0FQY6 A0A182I1G5 D3TQ17 A0A1A9XXP5 A0A182GVL7 A0A182V9W8 T1GXD3 A0A182JC26 A0A182FEU9 A0A1B0ADQ5 Q29HX1 A0A1A9UEL1
A0A3S2LZL4 A0A0L7LLF6 A0A212FP06 S4PSS1 A0A1Y1M004 A0A195F485 E2BSI7 E9J329 A0A195D9X0 A0A151IEY8 A0A026X013 A0A151XEI1 A0A0T6AWF3 A0A0J7NT61 A0A154PLX8 B4JKN1 A0A088A2P6 A0A2A3EH38 A0A0L7RB87 A0A0C9QXG6 A0A1S3D0W0 A0A0K8TP88 B4M1R6 A0A1W4X925 A0A139WER1 A0A0N0U5E8 C4WSW3 B4L8S4 A0A0M3QZC1 A0A1J1IBA0 B4NC55 A0A0K8VTR4 K7J5W9 A0A034V6N8 B3MZN3 A0A2S2QKY2 A0A067RF48 A0A1B0GLA7 A0A084WRL8 A0A1I8P3S3 A0A069DQH3 A0A3B0K7E0 B4H4N0 A0A0L0C4G1 A0A1B6CCP3 A0A2H8TPK8 J3JUM2 U5EWB0 A0A023EL58 A0A1W7R511 A0A1W4VHH0 A0A1L8D9H1 A0A224XYX6 E0VEV3 A0A1B6IG51 A0A1B6GF21 A0A0A9W7P8 A0A1I8MY21 A0A2M4AW38 Q16J55 M9NF00 Q9U4H5 B4PWU5 B4ILP3 B3NT89 A0A182XP59 A0A2S2P3F1 Q7Q602 A0A182TV90 B4NU72 T1J3U4 A0A182JZ66 A0A182WHV1 A0A182RGT1 W5J4D3 A0A182QWN4 A0A182NLQ7 A0A1S4GRB1 A0A182KP22 A0A182P108 A0A182LZQ8 A0A1B0FQY6 A0A182I1G5 D3TQ17 A0A1A9XXP5 A0A182GVL7 A0A182V9W8 T1GXD3 A0A182JC26 A0A182FEU9 A0A1B0ADQ5 Q29HX1 A0A1A9UEL1
Ontologies
GO
Topology
Subcellular location
Membrane
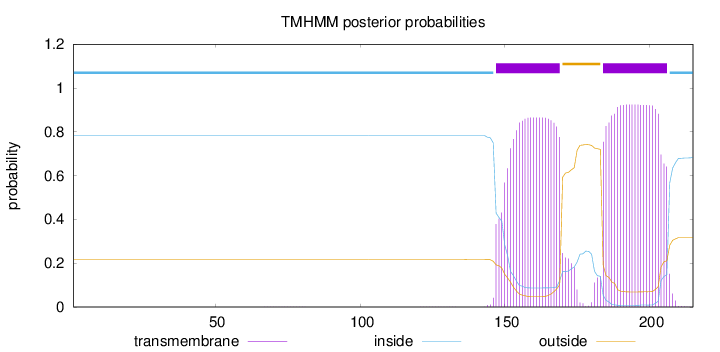
Length:
215
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
39.33841
Exp number, first 60 AAs:
0.0004
Total prob of N-in:
0.78346
inside
1 - 146
TMhelix
147 - 169
outside
170 - 183
TMhelix
184 - 206
inside
207 - 215
Population Genetic Test Statistics
Pi
278.714326
Theta
183.009878
Tajima's D
1.623053
CLR
51.584911
CSRT
0.813509324533773
Interpretation
Uncertain
