Gene
KWMTBOMO11585
Pre Gene Modal
BGIBMGA004097
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_probable_cytochrome_P450_49a1_[Bombyx_mori]
Full name
Probable cytochrome P450 49a1
Alternative Name
CYPXLIXA1
Location in the cell
Mitochondrial Reliability : 2.84
Sequence
CDS
ATGGACAAAGCTCGGTGTTACACAGTCATGCCGGGACCAAGGCCCCTGCCTATTCTCGGCAACTCATGGAGGTTCGCATTAGGCTGGAAACCATGGAGAACGAAAAGGCTGGACTTGACGCTTTGGTGTTTACGATCGTTAGCCGGCGCGGGCGGTGCCGCTAAAGTGGCCAAACTATTCGGTCATCCCGATCTCGTTTTCCCTTTCTGCGCCGAAGAAACCGCGAGGATATACAGGAGGGAAGATGCGATGCCCCACAGAGCCGCAGCTCCATGTCTGAAGCACTACAAACAAGAGTTAAGGAAAGATTTCTTCGGAGACGAACCAGGACTTATTGGAATTCACGGAGAACCGTGGTCAAGATTTCGGTCAAAAGTATCCAAAGCATTAATAGCTCCCGAAGCGGCACGTGCTGCTGTACCGGAGTTAGATTACGTCGCTAATGACTTTGTTATAAGATTAGAACATCTCCTTGATCTGAACAGGGAACTTCCAAAAGATTTCCTCACGGAGCTCTATAAATGGGCTCTCGAATCTGTGGGCGCGTGGGCTTTGGGCACACGATTAGGATGTTTAAACGATACCAAAACGGACGCTAAGGAAATGATAAAATGTATACATGGATTTTTTCACAGTGTACCCGAATTGGAGTTGTCCGCACCGTTATGGAGAATATATTCAACGCCCGCTTACAAAACCTATATCGAAGCACTGGACTCTTTCAGATTACTCTGTTTAAAAAGATTAACTGATAAAGGAGTCTGTGCTCAGGTGGCTAAAAATTGCGGTCAGAAAGTAGCCACAATCCTGGCTTTAGATTTAATGTTAGTTGGGGTGGACACAACAGCCGCCGCAGCTGCGAGCTCTTTATACTTATTAGCTAATGTACCTAGGGCACAGAGAGCGCTTCAAAAAGAACTAGACACGAATTTACCGAAAAATAGGATATTGAATGATAAAGACTTGGACAAACTTCCTTATCTGAAAGCTTGTATAAAAGAAGCGTTGCGAATGAAACCGGTAATTTTGGGAAACGGACGATGCATTCAATCGGACACCACCATATCTGGCTACAAAGTTCCAAAAGGCCTCGGACGAGATATCGAGAACATGCGATCGTACAGTAGTGTCGGACGTTTGTCGCAAGCAGAAGGAAATAGGGATACACCCGTTCGCGTCACTCCCCTTTGGATTTGGTCGACGGATGTGCGTCGGGAAGAGATTCGCCGAAGTCGAATTGCAACTTTTACTGGCCAGGGTAATCATTGA
Protein
MDKARCYTVMPGPRPLPILGNSWRFALGWKPWRTKRLDLTLWCLRSLAGAGGAAKVAKLFGHPDLVFPFCAEETARIYRREDAMPHRAAAPCLKHYKQELRKDFFGDEPGLIGIHGEPWSRFRSKVSKALIAPEAARAAVPELDYVANDFVIRLEHLLDLNRELPKDFLTELYKWALESVGAWALGTRLGCLNDTKTDAKEMIKCIHGFFHSVPELELSAPLWRIYSTPAYKTYIEALDSFRLLCLKRLTDKGVCAQVAKNCGQKVATILALDLMLVGVDTTAAAAASSLYLLANVPRAQRALQKELDTNLPKNRILNDKDLDKLPYLKACIKEALRMKPVILGNGRCIQSDTTISGYKVPKGLGRDIENMRSYSSVGRLSQAEGNRDTPVRVTPLWIWSTDVRREEIRRSRIATFTGQGNH
Summary
Description
May be involved in the metabolism of insect hormones and in the breakdown of synthetic insecticides.
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Keywords
Alternative splicing
Complete proteome
Endoplasmic reticulum
Heme
Iron
Membrane
Metal-binding
Microsome
Monooxygenase
Oxidoreductase
Reference proteome
Feature
chain Probable cytochrome P450 49a1
splice variant In isoform C.
splice variant In isoform C.
Uniprot
H9J3L0
A0A194QD08
A0A194QVQ2
A0A212EW64
A0A0L7LS76
A0A1B0G778
+ More
A0A1A9VGK8 A0A1A9X3H5 B3N6D8 B4KN43 A0A125S6N7 A0A0L0BP26 B4GG60 Q28XI1 B4HMQ3 A0A0B4K7M1 Q9V5L3 A0A3B0JFL4 A0A3B0JN88 B4NXN7 A0A0R1DMZ7 B4QIE0 B4N662 A0A068FC46 A0A1W4VCA1 A0A0M4ETM1 B4J7I9 B4LKE7 A0A0A1WFU7 W8BEW4 W8B9Y6 A0A0K8U800 A0A1I8MIW9 A0A1I8MIX8 A0A1I8QAX8 B3MI40 A0A182HWH4 A0A182XML5 A0A182VDU1 A0A182UA43 A0A182L361 Q7PNM9 A0A067R674 A0A1J1J2W8 A0A336JXF2 A0A182PLU9 A0A182FHV4 B0VZB4 A0A182Y248 A0A182QKV0 A0A1Q3FMD9 A0A182J2S6 A0A2M4BKS7 A0A2J7PDU4 W5J440 A0A182S4M1 Q16Y74 E0VDR0 D6WQ58 A0A0M4HGU6 J9JRR2 A0A1B6CMB7 A0A2D1GSD3 A0A2H8TLX1 A0A0K0LB65 A0A2S2QSX9 A0A1V0D9E8 A0A182JZ57 A0A067RSD6 I1VJ31 A0A182NJR1 A0A182SPT5 A0A182WKZ1 A0A248QEZ9 A0A194Q424 A0A0M9A7Y8 A0A3S2NTQ9 E2AFK1 A0A154PR32 A0A2A3EML1 A0A2W1C0B2 A0A026WVH3 A0A212FL05 A0A2J7PYK4 A0A088A457 A0A0M4H594 K7J2R1 A0A232F204 A0A0L7R6B8 A0A2J7PYM6 A0A2J7PYJ4 A0A2A4K848 D2A103 E0W2B1 A0A1J1J892 A0A023F3W6 A0A1W6L1F3
A0A1A9VGK8 A0A1A9X3H5 B3N6D8 B4KN43 A0A125S6N7 A0A0L0BP26 B4GG60 Q28XI1 B4HMQ3 A0A0B4K7M1 Q9V5L3 A0A3B0JFL4 A0A3B0JN88 B4NXN7 A0A0R1DMZ7 B4QIE0 B4N662 A0A068FC46 A0A1W4VCA1 A0A0M4ETM1 B4J7I9 B4LKE7 A0A0A1WFU7 W8BEW4 W8B9Y6 A0A0K8U800 A0A1I8MIW9 A0A1I8MIX8 A0A1I8QAX8 B3MI40 A0A182HWH4 A0A182XML5 A0A182VDU1 A0A182UA43 A0A182L361 Q7PNM9 A0A067R674 A0A1J1J2W8 A0A336JXF2 A0A182PLU9 A0A182FHV4 B0VZB4 A0A182Y248 A0A182QKV0 A0A1Q3FMD9 A0A182J2S6 A0A2M4BKS7 A0A2J7PDU4 W5J440 A0A182S4M1 Q16Y74 E0VDR0 D6WQ58 A0A0M4HGU6 J9JRR2 A0A1B6CMB7 A0A2D1GSD3 A0A2H8TLX1 A0A0K0LB65 A0A2S2QSX9 A0A1V0D9E8 A0A182JZ57 A0A067RSD6 I1VJ31 A0A182NJR1 A0A182SPT5 A0A182WKZ1 A0A248QEZ9 A0A194Q424 A0A0M9A7Y8 A0A3S2NTQ9 E2AFK1 A0A154PR32 A0A2A3EML1 A0A2W1C0B2 A0A026WVH3 A0A212FL05 A0A2J7PYK4 A0A088A457 A0A0M4H594 K7J2R1 A0A232F204 A0A0L7R6B8 A0A2J7PYM6 A0A2J7PYJ4 A0A2A4K848 D2A103 E0W2B1 A0A1J1J892 A0A023F3W6 A0A1W6L1F3
EC Number
1.14.-.-
Pubmed
19121390
26354079
22118469
26227816
17994087
18057021
+ More
26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 17550304 22936249 25880816 25830018 24495485 25315136 20966253 12364791 14747013 17210077 24845553 25244985 20920257 23761445 17510324 20566863 18362917 19820115 26319543 22516182 28881445 20798317 28756777 24508170 30249741 20075255 28648823 25474469
26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 17550304 22936249 25880816 25830018 24495485 25315136 20966253 12364791 14747013 17210077 24845553 25244985 20920257 23761445 17510324 20566863 18362917 19820115 26319543 22516182 28881445 20798317 28756777 24508170 30249741 20075255 28648823 25474469
EMBL
BABH01021564
BABH01021565
KQ459185
KPJ03304.1
KQ461073
KPJ09613.1
+ More
AGBW02012080 OWR45707.1 JTDY01000218 KOB78234.1 CCAG010008069 CH954177 EDV58107.1 KQS70245.1 CH933808 EDW08870.1 KRG04326.1 KRG04327.1 KT601129 AME15798.1 JRES01001578 KNC21835.1 CH479183 EDW35480.1 CM000071 EAL26335.1 CH480816 EDW47265.1 AE013599 AFH08010.1 AY075453 AY102688 BT056244 OUUW01000001 SPP74110.1 SPP74111.1 CM000157 EDW89662.1 KRJ98649.1 KRJ98650.1 CM000362 CM002911 EDX06500.1 KMY92793.1 CH964154 EDW79851.2 KJ702277 AID61431.1 CP012524 ALC40607.1 CH916367 EDW01113.1 CH940648 EDW61738.1 GBXI01016535 JAC97756.1 GAMC01011087 JAB95468.1 GAMC01011083 JAB95472.1 GDHF01029542 JAI22772.1 CH902619 EDV38050.1 KPU77234.1 APCN01001491 AAAB01008960 EAA11902.5 KK852927 KDR13690.1 CVRI01000067 CRL06719.1 UFQS01000004 UFQT01000004 SSW96881.1 SSX17268.1 DS231813 EDS25689.1 AXCN02000515 AXCN02000516 GFDL01006328 JAV28717.1 GGFJ01004535 MBW53676.1 NEVH01026142 PNF14497.1 ADMH02002165 ETN58108.1 CH477523 EAT39564.1 DS235083 EEB11516.1 KQ971354 EFA07521.1 KR012817 ALD15892.1 ABLF02025275 ABLF02025279 ABLF02046672 GEDC01022689 JAS14609.1 MF471397 ATN96003.1 GFXV01002483 MBW14288.1 KM216993 AIW79956.1 GGMS01011631 MBY80834.1 KY212061 ARA91625.1 KK852446 KDR23725.1 JQ855652 AFI45015.1 KX443475 ASO98050.1 KQ459585 KPI98160.1 KQ435728 KOX77763.1 RSAL01000206 RVE44347.1 GL439118 EFN67752.1 KQ435066 KZC14341.1 KZ288219 PBC32281.1 KZ149927 PZC77503.1 KK107085 QOIP01000008 EZA60040.1 RLU19440.1 AGBW02007925 OWR54417.1 NEVH01020348 PNF21416.1 KR012815 ALD15890.1 NNAY01001258 OXU24592.1 KQ414648 KOC66321.1 PNF21412.1 PNF21417.1 NWSH01000071 PCG79963.1 KQ971338 EFA02807.1 DS235875 EEB19767.1 CVRI01000075 CRL08607.1 GBBI01002577 JAC16135.1 KX609544 ARN17951.1
AGBW02012080 OWR45707.1 JTDY01000218 KOB78234.1 CCAG010008069 CH954177 EDV58107.1 KQS70245.1 CH933808 EDW08870.1 KRG04326.1 KRG04327.1 KT601129 AME15798.1 JRES01001578 KNC21835.1 CH479183 EDW35480.1 CM000071 EAL26335.1 CH480816 EDW47265.1 AE013599 AFH08010.1 AY075453 AY102688 BT056244 OUUW01000001 SPP74110.1 SPP74111.1 CM000157 EDW89662.1 KRJ98649.1 KRJ98650.1 CM000362 CM002911 EDX06500.1 KMY92793.1 CH964154 EDW79851.2 KJ702277 AID61431.1 CP012524 ALC40607.1 CH916367 EDW01113.1 CH940648 EDW61738.1 GBXI01016535 JAC97756.1 GAMC01011087 JAB95468.1 GAMC01011083 JAB95472.1 GDHF01029542 JAI22772.1 CH902619 EDV38050.1 KPU77234.1 APCN01001491 AAAB01008960 EAA11902.5 KK852927 KDR13690.1 CVRI01000067 CRL06719.1 UFQS01000004 UFQT01000004 SSW96881.1 SSX17268.1 DS231813 EDS25689.1 AXCN02000515 AXCN02000516 GFDL01006328 JAV28717.1 GGFJ01004535 MBW53676.1 NEVH01026142 PNF14497.1 ADMH02002165 ETN58108.1 CH477523 EAT39564.1 DS235083 EEB11516.1 KQ971354 EFA07521.1 KR012817 ALD15892.1 ABLF02025275 ABLF02025279 ABLF02046672 GEDC01022689 JAS14609.1 MF471397 ATN96003.1 GFXV01002483 MBW14288.1 KM216993 AIW79956.1 GGMS01011631 MBY80834.1 KY212061 ARA91625.1 KK852446 KDR23725.1 JQ855652 AFI45015.1 KX443475 ASO98050.1 KQ459585 KPI98160.1 KQ435728 KOX77763.1 RSAL01000206 RVE44347.1 GL439118 EFN67752.1 KQ435066 KZC14341.1 KZ288219 PBC32281.1 KZ149927 PZC77503.1 KK107085 QOIP01000008 EZA60040.1 RLU19440.1 AGBW02007925 OWR54417.1 NEVH01020348 PNF21416.1 KR012815 ALD15890.1 NNAY01001258 OXU24592.1 KQ414648 KOC66321.1 PNF21412.1 PNF21417.1 NWSH01000071 PCG79963.1 KQ971338 EFA02807.1 DS235875 EEB19767.1 CVRI01000075 CRL08607.1 GBBI01002577 JAC16135.1 KX609544 ARN17951.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000092444
+ More
UP000078200 UP000091820 UP000008711 UP000009192 UP000037069 UP000008744 UP000001819 UP000001292 UP000000803 UP000268350 UP000002282 UP000000304 UP000007798 UP000192221 UP000092553 UP000001070 UP000008792 UP000095301 UP000095300 UP000007801 UP000075840 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000027135 UP000183832 UP000075885 UP000069272 UP000002320 UP000076408 UP000075886 UP000075880 UP000235965 UP000000673 UP000075900 UP000008820 UP000009046 UP000007266 UP000007819 UP000075881 UP000075884 UP000075901 UP000075920 UP000053105 UP000283053 UP000000311 UP000076502 UP000242457 UP000053097 UP000279307 UP000005203 UP000002358 UP000215335 UP000053825 UP000218220
UP000078200 UP000091820 UP000008711 UP000009192 UP000037069 UP000008744 UP000001819 UP000001292 UP000000803 UP000268350 UP000002282 UP000000304 UP000007798 UP000192221 UP000092553 UP000001070 UP000008792 UP000095301 UP000095300 UP000007801 UP000075840 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000027135 UP000183832 UP000075885 UP000069272 UP000002320 UP000076408 UP000075886 UP000075880 UP000235965 UP000000673 UP000075900 UP000008820 UP000009046 UP000007266 UP000007819 UP000075881 UP000075884 UP000075901 UP000075920 UP000053105 UP000283053 UP000000311 UP000076502 UP000242457 UP000053097 UP000279307 UP000005203 UP000002358 UP000215335 UP000053825 UP000218220
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9J3L0
A0A194QD08
A0A194QVQ2
A0A212EW64
A0A0L7LS76
A0A1B0G778
+ More
A0A1A9VGK8 A0A1A9X3H5 B3N6D8 B4KN43 A0A125S6N7 A0A0L0BP26 B4GG60 Q28XI1 B4HMQ3 A0A0B4K7M1 Q9V5L3 A0A3B0JFL4 A0A3B0JN88 B4NXN7 A0A0R1DMZ7 B4QIE0 B4N662 A0A068FC46 A0A1W4VCA1 A0A0M4ETM1 B4J7I9 B4LKE7 A0A0A1WFU7 W8BEW4 W8B9Y6 A0A0K8U800 A0A1I8MIW9 A0A1I8MIX8 A0A1I8QAX8 B3MI40 A0A182HWH4 A0A182XML5 A0A182VDU1 A0A182UA43 A0A182L361 Q7PNM9 A0A067R674 A0A1J1J2W8 A0A336JXF2 A0A182PLU9 A0A182FHV4 B0VZB4 A0A182Y248 A0A182QKV0 A0A1Q3FMD9 A0A182J2S6 A0A2M4BKS7 A0A2J7PDU4 W5J440 A0A182S4M1 Q16Y74 E0VDR0 D6WQ58 A0A0M4HGU6 J9JRR2 A0A1B6CMB7 A0A2D1GSD3 A0A2H8TLX1 A0A0K0LB65 A0A2S2QSX9 A0A1V0D9E8 A0A182JZ57 A0A067RSD6 I1VJ31 A0A182NJR1 A0A182SPT5 A0A182WKZ1 A0A248QEZ9 A0A194Q424 A0A0M9A7Y8 A0A3S2NTQ9 E2AFK1 A0A154PR32 A0A2A3EML1 A0A2W1C0B2 A0A026WVH3 A0A212FL05 A0A2J7PYK4 A0A088A457 A0A0M4H594 K7J2R1 A0A232F204 A0A0L7R6B8 A0A2J7PYM6 A0A2J7PYJ4 A0A2A4K848 D2A103 E0W2B1 A0A1J1J892 A0A023F3W6 A0A1W6L1F3
A0A1A9VGK8 A0A1A9X3H5 B3N6D8 B4KN43 A0A125S6N7 A0A0L0BP26 B4GG60 Q28XI1 B4HMQ3 A0A0B4K7M1 Q9V5L3 A0A3B0JFL4 A0A3B0JN88 B4NXN7 A0A0R1DMZ7 B4QIE0 B4N662 A0A068FC46 A0A1W4VCA1 A0A0M4ETM1 B4J7I9 B4LKE7 A0A0A1WFU7 W8BEW4 W8B9Y6 A0A0K8U800 A0A1I8MIW9 A0A1I8MIX8 A0A1I8QAX8 B3MI40 A0A182HWH4 A0A182XML5 A0A182VDU1 A0A182UA43 A0A182L361 Q7PNM9 A0A067R674 A0A1J1J2W8 A0A336JXF2 A0A182PLU9 A0A182FHV4 B0VZB4 A0A182Y248 A0A182QKV0 A0A1Q3FMD9 A0A182J2S6 A0A2M4BKS7 A0A2J7PDU4 W5J440 A0A182S4M1 Q16Y74 E0VDR0 D6WQ58 A0A0M4HGU6 J9JRR2 A0A1B6CMB7 A0A2D1GSD3 A0A2H8TLX1 A0A0K0LB65 A0A2S2QSX9 A0A1V0D9E8 A0A182JZ57 A0A067RSD6 I1VJ31 A0A182NJR1 A0A182SPT5 A0A182WKZ1 A0A248QEZ9 A0A194Q424 A0A0M9A7Y8 A0A3S2NTQ9 E2AFK1 A0A154PR32 A0A2A3EML1 A0A2W1C0B2 A0A026WVH3 A0A212FL05 A0A2J7PYK4 A0A088A457 A0A0M4H594 K7J2R1 A0A232F204 A0A0L7R6B8 A0A2J7PYM6 A0A2J7PYJ4 A0A2A4K848 D2A103 E0W2B1 A0A1J1J892 A0A023F3W6 A0A1W6L1F3
PDB
3K9Y
E-value=2.42355e-21,
Score=252
Ontologies
KEGG
GO
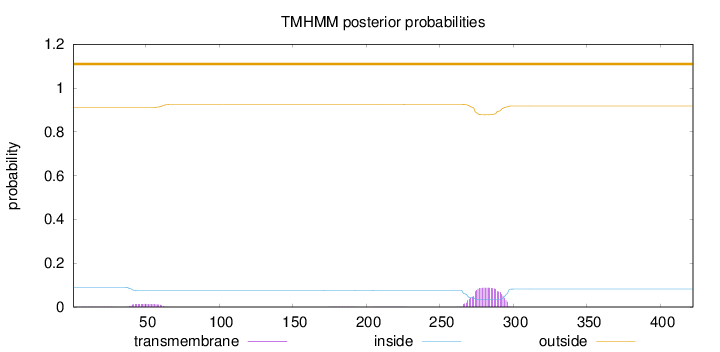
Topology
Subcellular location
Endoplasmic reticulum membrane
Microsome membrane
Microsome membrane
Length:
422
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.23122
Exp number, first 60 AAs:
0.26463
Total prob of N-in:
0.08904
outside
1 - 422
Population Genetic Test Statistics
Pi
204.534955
Theta
149.728948
Tajima's D
0.388539
CLR
0.000732
CSRT
0.47847607619619
Interpretation
Uncertain
